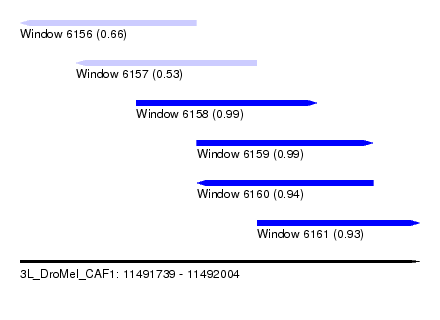
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 11,491,739 – 11,492,004 |
| Length | 265 |
| Max. P | 0.992598 |

| Location | 11,491,739 – 11,491,856 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.97 |
| Mean single sequence MFE | -43.57 |
| Consensus MFE | -30.92 |
| Energy contribution | -31.07 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.661249 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11491739 117 - 23771897 CCCGGUAAUGCCAACGGUUGACUGGCUGCCGCCAGAAAGCUGGGCAACAGCACCUGUCGCUGGUACAAAUCGAUCAGCUGCUUAAACUGCAACAGAUUCAUAUUCAGAGCUGGUCCC--- .(((((..((((..(((((..(((((....)))))..)))))))))((((...)))).)))))........((((((((((.......))....((.......))..))))))))..--- ( -38.30) >DroPse_CAF1 13066 112 - 1 --------CGGCAUCGGCUGGCUGGCCGCGGCCAGGAAGCUGGGCAGGAGCACCUGCCUCUGGUACAGGUCGAUCAGCUGUUUGAACUGCAGCAGAUUCAGAUUCAGACCCGAUUCCGAU --------(((.((((((((..(((((...(((((......(((((((....))))))))))))...)))))..)))).((((((((((.(.....).))).))))))).)))).))).. ( -50.60) >DroSec_CAF1 6806 117 - 1 CCCGGUAAUGCCAACGGUUGACUGGCUGCGGCCAAAAAGCUGGGCAACAGCACCUGUCGCUGGUACAAAUCGAUCAGCUGCUUAAACUGCAACAGAUUCAUAUUCAGAGCUGGUCCC--- .(((((..((((..(((((...((((....))))...)))))))))((((...)))).)))))........((((((((((.......))....((.......))..))))))))..--- ( -35.40) >DroPer_CAF1 3286 107 - 1 --------CGGCAUCGGCUGGCUGGCCGCGGCCAGGAAGCUGGGCAGGAGCACCUGCCUCUGGUACAGGUCGAUCAGCUGUUUGAACUGCAGCAGAUUCAGAUUCAGACCCGAUC----- --------(((....(((((..(((((...(((((......(((((((....))))))))))))...)))))..)))))((((((((((.(.....).))).))))))))))...----- ( -46.30) >DroEre_CAF1 2493 117 - 1 CCCGGUAACGCCAGUGGUUGGCUGGCUGCCGCCAGAAAGCUGGGCAGCAGCACCUGUCGCUGGUACAAAUCGAUCAGCUGCUUAAACUGCAGCAGAUUCAUAUUCAAAGCUGGUCCC--- .(((((...((((((((..((...(((((.(((((....)).))).))))).))..)))))))).......((((.(((((.......))))).))))..........)))))....--- ( -47.20) >DroYak_CAF1 3165 117 - 1 CCCGGCAACGCCAACGGUUGACUGGCUGCCGCAAGAAAGCUGGGCAGCAGCACCUGCCGCUGGUACAAAUCGAUCAGCUGCUUGAACUGCAGCAGAUUCAUAUUCAACGCUGGUCCC--- .(((((...((((.((((((.(((.(((((((......))..))))))))))...)))).)))).......((((.(((((.......))))).))))..........)))))....--- ( -43.60) >consensus CCCGGUAACGCCAACGGUUGACUGGCUGCCGCCAGAAAGCUGGGCAGCAGCACCUGCCGCUGGUACAAAUCGAUCAGCUGCUUAAACUGCAGCAGAUUCAUAUUCAGAGCUGGUCCC___ .........((((.(((((..(((((....)))))..)))))(((((......)))))..)))).......((((.(((((.......))))).((.......))......))))..... (-30.92 = -31.07 + 0.14)

| Location | 11,491,776 – 11,491,896 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.17 |
| Mean single sequence MFE | -46.06 |
| Consensus MFE | -38.78 |
| Energy contribution | -40.18 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.84 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.525656 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11491776 120 - 23771897 GUCCAGAAGAGGCCCUGAAACUCCAGGACCUGCUGCUGAACCCGGUAAUGCCAACGGUUGACUGGCUGCCGCCAGAAAGCUGGGCAACAGCACCUGUCGCUGGUACAAAUCGAUCAGCUG .......(((((.((((......)))).))).))(((((...((((..((((..(((((..(((((....)))))..)))))(((.((((...)))).)))))))...)))).))))).. ( -42.80) >DroSec_CAF1 6843 120 - 1 GUCCAGAAGGGAUCCUGAAGCUCCAGGACCUGCUGCUGAACCCGGUAAUGCCAACGGUUGACUGGCUGCGGCCAAAAAGCUGGGCAACAGCACCUGUCGCUGGUACAAAUCGAUCAGCUG ((((((..(((.(((((......))))))))(((((.....(((((...(((...)))..)))))..))))).......)))))).((((...)))).((((((........)))))).. ( -43.10) >DroSim_CAF1 740 120 - 1 GUCCAGAAGGGAUCCUGAAGCUCCAGGACCUGCUGCUGAGCCCGGUAAUGCCAACGGUUGACUGGCUGCCGCCAGGAAGCUGGGCAACAGCACCUGUCGCUGGUACAAAUCGAUCAGCUG ...(((..(((.(((((......))))))))..(((((.((((((.....))...((((..(((((....)))))..))))))))..))))).)))..((((((........)))))).. ( -48.80) >DroEre_CAF1 2530 120 - 1 GUCCAGCAGGGGUCCUGAAACUGCGGGACCUGCUCCUGAUCCCGGUAACGCCAGUGGUUGGCUGGCUGCCGCCAGAAAGCUGGGCAGCAGCACCUGUCGCUGGUACAAAUCGAUCAGCUG .....((((((((((((......)))))))((((.(((....(((((..((((((.....)))))))))))((((....)))).))).))))))))).((((((........)))))).. ( -52.80) >DroYak_CAF1 3202 120 - 1 GUCCAGAAGUGGUCCCGACACUGCAGGACCUGCUCCUGAACCCGGCAACGCCAACGGUUGACUGGCUGCCGCAAGAAAGCUGGGCAGCAGCACCUGCCGCUGGUACAAAUCGAUCAGCUG ((.(((.((((((((.(......).))))).))).))).))..((((..((...(((....)))((((((((......))..)))))).))...))))((((((........)))))).. ( -42.80) >consensus GUCCAGAAGGGGUCCUGAAACUCCAGGACCUGCUGCUGAACCCGGUAAUGCCAACGGUUGACUGGCUGCCGCCAGAAAGCUGGGCAACAGCACCUGUCGCUGGUACAAAUCGAUCAGCUG ...(((...((((((((......))))))))((((.((..((((((...)))...((((..(((((....)))))..))))))))).))))..)))..((((((........)))))).. (-38.78 = -40.18 + 1.40)


| Location | 11,491,816 – 11,491,936 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.50 |
| Mean single sequence MFE | -48.68 |
| Consensus MFE | -41.04 |
| Energy contribution | -41.08 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.33 |
| SVM RNA-class probability | 0.992535 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11491816 120 + 23771897 GCUUUCUGGCGGCAGCCAGUCAACCGUUGGCAUUACCGGGUUCAGCAGCAGGUCCUGGAGUUUCAGGGCCUCUUCUGGACAUGCGCAUUUUGCUUGAGAAUGCCGCUCAACAGAAGCUAA ((((((((((....)))))......((((((........((((((.((.(((((((((....))))))))))).))))))..(.(((((((.....)))))))))).)))).)))))... ( -49.20) >DroSec_CAF1 6883 120 + 1 GCUUUUUGGCCGCAGCCAGUCAACCGUUGGCAUUACCGGGUUCAGCAGCAGGUCCUGGAGCUUCAGGAUCCCUUCUGGACAUGCGCAUUUUGCUGGAGAAUGCCGCUCAACAGAAGCUAA ((((((((((....)))).......((((((........((((((.((..((((((((....)))))))).)).))))))..(.(((((((.....)))))))))).))))))))))... ( -41.40) >DroSim_CAF1 780 120 + 1 GCUUCCUGGCGGCAGCCAGUCAACCGUUGGCAUUACCGGGCUCAGCAGCAGGUCCUGGAGCUUCAGGAUCCCUUCUGGACAUGCGCAUUUUGCUGGAGAAUGCCGCUCAACAGAAGCUAA (((((.((((((((.((((.(((....((((((..(((((((....))).((((((((....))))))))....))))..)))).))..)))))))....)))))).))...)))))... ( -48.20) >DroEre_CAF1 2570 120 + 1 GCUUUCUGGCGGCAGCCAGCCAACCACUGGCGUUACCGGGAUCAGGAGCAGGUCCCGCAGUUUCAGGACCCCUGCUGGACAUGCGACUCUUGCUGGAGAAUGCCGCUCAACAAAAGCUAA (((((.(((((((((((((.......)))))....((((.(..((..(((.((((.((((...........)))).)))).)))..))..).))))....)))))).))...)))))... ( -50.00) >DroYak_CAF1 3242 120 + 1 GCUUUCUUGCGGCAGCCAGUCAACCGUUGGCGUUGCCGGGUUCAGGAGCAGGUCCUGCAGUGUCGGGACCACUUCUGGACAUGCGACUCCUGCUGGAGAAUGCCGCUCAACAGAAGCUAA (((((...((((((.(((((........((.(((((...(((((((((..(((((((......))))))).)))))))))..))))).)).)))))....))))))......)))))... ( -54.60) >consensus GCUUUCUGGCGGCAGCCAGUCAACCGUUGGCAUUACCGGGUUCAGCAGCAGGUCCUGGAGUUUCAGGACCCCUUCUGGACAUGCGCAUUUUGCUGGAGAAUGCCGCUCAACAGAAGCUAA (((((..(((((((.(((((......((((.....))))((((((.((..(((((((......))))))).)).))))))...........)))))....))))))).....)))))... (-41.04 = -41.08 + 0.04)

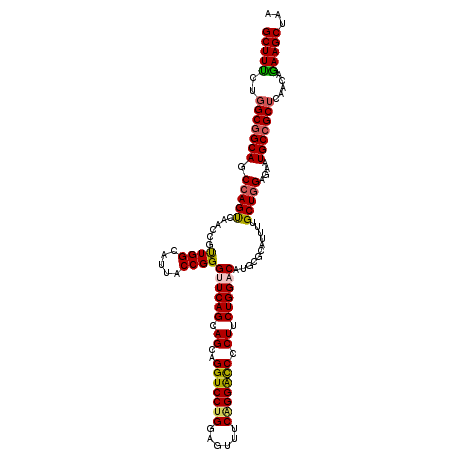
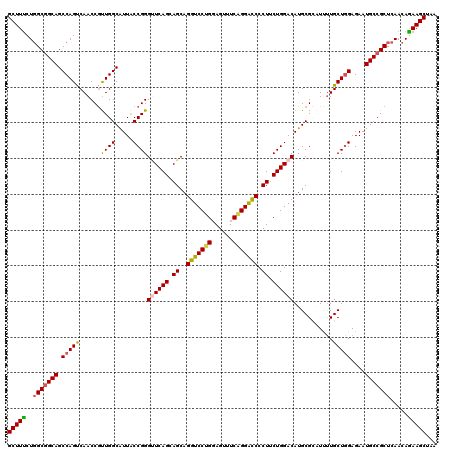
| Location | 11,491,856 – 11,491,973 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 90.85 |
| Mean single sequence MFE | -44.66 |
| Consensus MFE | -37.10 |
| Energy contribution | -38.18 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.34 |
| SVM RNA-class probability | 0.992598 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11491856 117 + 23771897 UUCAGCAGCAGGUCCUGGAGUUUCAGGGCCUCUUCUGGACAUGCGCAUUUUGCUUGAGAAUGCCGCUCAACAGAAGCUAAUGCUGCUCAAUGCAGCCGCAAGUGCAACGGCAAGUGC ...(((((((((((((((....)))))))).((((((.....(((((((((.....)))))).)))....))))))....)))))))..........(((..(((....)))..))) ( -45.80) >DroSec_CAF1 6923 117 + 1 UUCAGCAGCAGGUCCUGGAGCUUCAGGAUCCCUUCUGGACAUGCGCAUUUUGCUGGAGAAUGCCGCUCAACAGAAGCUAAUGCUGCUCAAUGCAGCUGCAAGUGCAACGGCAAGUGC ...(((((((((((((((....)))))))).((((((.....(((((((((.....)))))).)))....))))))....)))))))..........(((..(((....)))..))) ( -43.30) >DroSim_CAF1 820 117 + 1 CUCAGCAGCAGGUCCUGGAGCUUCAGGAUCCCUUCUGGACAUGCGCAUUUUGCUGGAGAAUGCCGCUCAACAGAAGCUAAUGCUGCUGAAUGCAGCUGCAAGUGCAACGGCAAGUGC .(((((((((((((((((....)))))))).((((((.....(((((((((.....)))))).)))....))))))....)))))))))........(((..(((....)))..))) ( -48.30) >DroEre_CAF1 2610 117 + 1 AUCAGGAGCAGGUCCCGCAGUUUCAGGACCCCUGCUGGACAUGCGACUCUUGCUGGAGAAUGCCGCUCAACAAAAGCUAAUGCUCCUCAAUGCAGCGGCAAGUGCAACGGCAAGUGC ...((..(((.((((.((((...........)))).)))).)))..))(((((((..(..(((((((((.....(((....)))......)).)))))))....)..)))))))... ( -42.80) >DroYak_CAF1 3282 117 + 1 UUCAGGAGCAGGUCCUGCAGUGUCGGGACCACUUCUGGACAUGCGACUCCUGCUGGAGAAUGCCGCUCAACAGAAGCUAAUGCUGCUCAAUGCAGCUGCAAGUGCAACAGCAAGUGC ......(((.(((((((......)))))))..(((((.....(((.((((....)))).....)))....))))))))...(((((.....))))).(((..(((....)))..))) ( -43.10) >consensus UUCAGCAGCAGGUCCUGGAGUUUCAGGACCCCUUCUGGACAUGCGCAUUUUGCUGGAGAAUGCCGCUCAACAGAAGCUAAUGCUGCUCAAUGCAGCUGCAAGUGCAACGGCAAGUGC ...((((((((((((((......))))))).((((((.....(((((((((.....)))))).)))....))))))....)))))))..........(((..(((....)))..))) (-37.10 = -38.18 + 1.08)


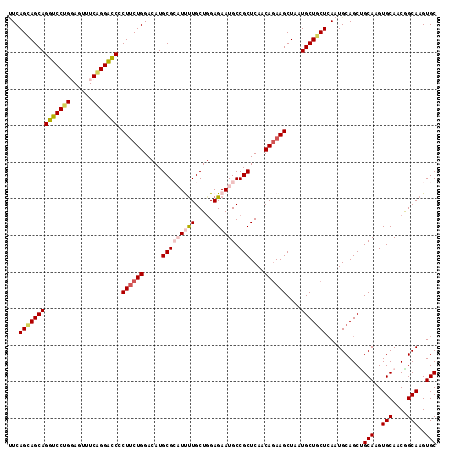
| Location | 11,491,856 – 11,491,973 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 90.85 |
| Mean single sequence MFE | -43.62 |
| Consensus MFE | -35.52 |
| Energy contribution | -36.36 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.939967 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11491856 117 - 23771897 GCACUUGCCGUUGCACUUGCGGCUGCAUUGAGCAGCAUUAGCUUCUGUUGAGCGGCAUUCUCAAGCAAAAUGCGCAUGUCCAGAAGAGGCCCUGAAACUCCAGGACCUGCUGCUGAA (((...(((((.......)))))))).((.((((((.....((((((....(((.((((.........))))))).....))))))(((.((((......)))).))))))))).)) ( -41.80) >DroSec_CAF1 6923 117 - 1 GCACUUGCCGUUGCACUUGCAGCUGCAUUGAGCAGCAUUAGCUUCUGUUGAGCGGCAUUCUCCAGCAAAAUGCGCAUGUCCAGAAGGGAUCCUGAAGCUCCAGGACCUGCUGCUGAA .....(((.(((((....))))).)))((.(((((((....((((((....(((.((((.........))))))).....))))))((.(((((......)))))))))))))).)) ( -42.50) >DroSim_CAF1 820 117 - 1 GCACUUGCCGUUGCACUUGCAGCUGCAUUCAGCAGCAUUAGCUUCUGUUGAGCGGCAUUCUCCAGCAAAAUGCGCAUGUCCAGAAGGGAUCCUGAAGCUCCAGGACCUGCUGCUGAG .....(((.(((((....))))).))).(((((((((....((((((....(((.((((.........))))))).....))))))((.(((((......)))))))))))))))). ( -46.40) >DroEre_CAF1 2610 117 - 1 GCACUUGCCGUUGCACUUGCCGCUGCAUUGAGGAGCAUUAGCUUUUGUUGAGCGGCAUUCUCCAGCAAGAGUCGCAUGUCCAGCAGGGGUCCUGAAACUGCGGGACCUGCUCCUGAU ((((((...((((....(((((((.((...((((((....))))))..))))))))).....))))..)))).)).....(((.((((((((((......)))))))).)).))).. ( -44.60) >DroYak_CAF1 3282 117 - 1 GCACUUGCUGUUGCACUUGCAGCUGCAUUGAGCAGCAUUAGCUUCUGUUGAGCGGCAUUCUCCAGCAGGAGUCGCAUGUCCAGAAGUGGUCCCGACACUGCAGGACCUGCUCCUGAA (((..(((....)))..))).(((((.....)))))....((((((((((((.......)).)))))))))).(((.((((...((((.......))))...)))).)))....... ( -42.80) >consensus GCACUUGCCGUUGCACUUGCAGCUGCAUUGAGCAGCAUUAGCUUCUGUUGAGCGGCAUUCUCCAGCAAAAUGCGCAUGUCCAGAAGGGGUCCUGAAACUCCAGGACCUGCUGCUGAA .....(((.(((((....))))).)))...((((((.....((((((....(((((........))......))).....))))))((((((((......))))))))))))))... (-35.52 = -36.36 + 0.84)


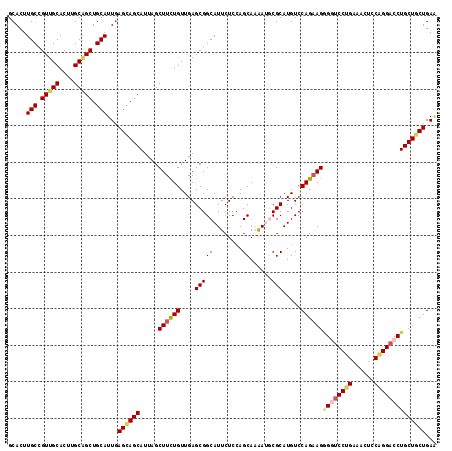
| Location | 11,491,896 – 11,492,004 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 87.10 |
| Mean single sequence MFE | -37.06 |
| Consensus MFE | -30.20 |
| Energy contribution | -31.12 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.931733 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11491896 108 + 23771897 AUGCGCAUUUUGCUUGAGAAUGCCGCUCAACAGAAGCUAAUGCUGCUCAAUGCAGCCGCAAGUGCAACGGCAAGUGCAGUGGCAGC---------CACAACAAUCAACUCCGGUCAA .(((((.(((((.(((((.......))))))))))(((...(((((.....))))).((....))...)))..)))))((((...)---------)))................... ( -31.00) >DroSec_CAF1 6963 108 + 1 AUGCGCAUUUUGCUGGAGAAUGCCGCUCAACAGAAGCUAAUGCUGCUCAAUGCAGCUGCAAGUGCAACGGCAAGUGCAGUGGCAGC---------CACAACAAUGGACUCCGGUCAA ...........(((((((..((((((...............(((((.....)))))((((..(((....)))..)))))))))).(---------((......))).)))))))... ( -39.70) >DroSim_CAF1 860 108 + 1 AUGCGCAUUUUGCUGGAGAAUGCCGCUCAACAGAAGCUAAUGCUGCUGAAUGCAGCUGCAAGUGCAACGGCAAGUGCAGUGGCAGC---------CACAACAAUGGACUAAGGUCAA .....(((((((.(((....((((((....(((.(((....))).)))...)).((((((..(((....)))..)))))))))).)---------))))).))))(((....))).. ( -38.20) >DroEre_CAF1 2650 117 + 1 AUGCGACUCUUGCUGGAGAAUGCCGCUCAACAAAAGCUAAUGCUCCUCAAUGCAGCGGCAAGUGCAACGGCAAGUGCGGUGGCAAGUGCGGCAACCACAACGAUGAAUUCCGGCCAG ...((.(.(((((((..(..(((((((((.....(((....)))......)).)))))))....)..))))))).))).((((..(((.(....))))..((........)))))). ( -39.80) >DroYak_CAF1 3322 108 + 1 AUGCGACUCCUGCUGGAGAAUGCCGCUCAACAGAAGCUAAUGCUGCUCAAUGCAGCUGCAAGUGCAACAGCAAGUGCCGUGGCAAG---------CACAACCAUGGACUCCGGGCAA .(((..((((....))))..((((((...............(((((.....))))).(((..(((....)))..))).)))))).)---------))...((.(((...)))))... ( -36.60) >consensus AUGCGCAUUUUGCUGGAGAAUGCCGCUCAACAGAAGCUAAUGCUGCUCAAUGCAGCUGCAAGUGCAACGGCAAGUGCAGUGGCAGC_________CACAACAAUGGACUCCGGUCAA ..........((((((((..(((((((..............(((((.....))))).(((..(((....)))..)))))))))).............((....))..)))))).)). (-30.20 = -31.12 + 0.92)

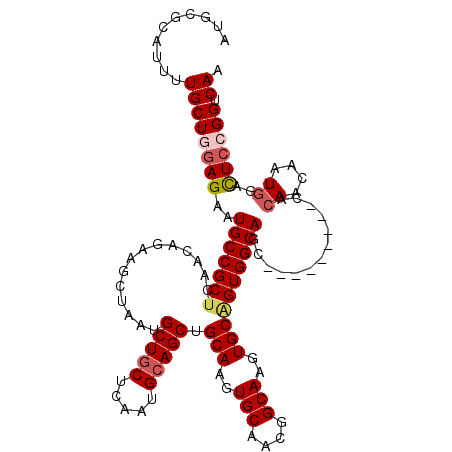
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:10:56 2006