| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 11,491,008 – 11,491,102 |
| Length | 94 |
| Max. P | 0.791452 |

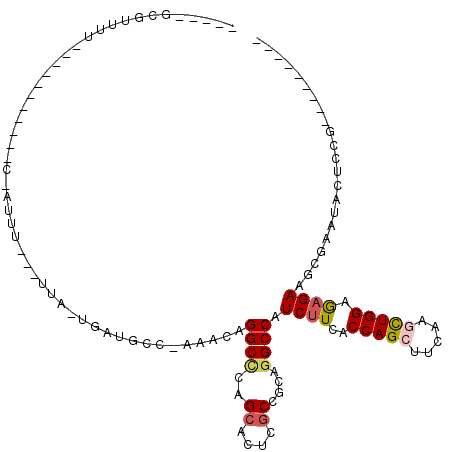
| Location | 11,491,008 – 11,491,102 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 67.26 |
| Mean single sequence MFE | -25.23 |
| Consensus MFE | -13.43 |
| Energy contribution | -13.65 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.791452 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11491008 94 + 23771897 ----CGCUUUUU-U--------UCAAUUU---UUA-UGCUUCCAAAAUAGGCCCAGCACUCGCCGCAGGCCAUCUUCACCAGCUUUAAGCUGGAGAGAAGCGAAUAUUCCG--------- ----((((((((-(--------.......---...-.............((((..((.......)).)))).......(((((.....)))))))))))))).........--------- ( -26.50) >DroPse_CAF1 12291 93 + 1 ----AGACC-----------------UCU---UUC-UGUUGCC--CGCAGGCUCAGCAUUCGCCCCAGGCCAUCUUCACCAGCUUCAAAUUGGAAAGAAAUGAGCACUCAAGUGCGCCUG ----.....-----------------...---..(-((..(((--....))).))).........(((((..((((..((((.......)))).)))).....((((....))))))))) ( -22.30) >DroSec_CAF1 6068 101 + 1 UGUACGCGUUUU-U---------CGAUUUAUUUUAUUUAUCCCAAAACAGGCCCAGCACUCACCGCAGGCCAUCUUCACCAGCUUUAAGCUGGAGAGAAGCGAAUACGCCG--------- .(((..((((((-(---------(.........................((((..((.......)).)))).......(((((.....))))).))))))))..)))....--------- ( -30.10) >DroPer_CAF1 2528 93 + 1 ----AGACC-----------------UCU---UUC-UGUUGCC--AGCAGGCUCAGCAUUCGCCCCAGGCCAUCUUCACCAGCUUUAAAUUGGAAAGAAAUGAGCACUCAAGUGCGCCUG ----.....-----------------...---..(-((..(((--....))).))).........(((((..((((..((((.......)))).)))).....((((....))))))))) ( -22.80) >DroEre_CAF1 1732 95 + 1 UAU--GCGUUUU----------UCUAUUU---UUA-UGAUGCCCAUACAGGCGCAGCACUCGCCGCAGGCCAUCUUCACCAGCUUCAAGCUGGAGCGAAGCGAAUACUCCG--------- ...--.((((((----------.((....---...-.((((.((.....((((.......))))...)).))))....(((((.....))))))).)))))).........--------- ( -25.80) >DroYak_CAF1 2322 105 + 1 UAU--ACGUUUUUUCCAUUUUUUUUUUUU---UUA-CGAUCCCCCCACAGGCCCAGCACUCGCCGCAGGCCAUCUUCACCAGCUUCAAGCUGGAGAGAAGCGAAUACUCUG--------- ...--.((((((((...............---...-.............((((..((.......)).)))).......(((((.....))))).)))))))).........--------- ( -23.90) >consensus _____GCGUUUU___________C_AUUU___UUA_UGAUGCC_AAACAGGCCCAGCACUCGCCGCAGGCCAUCUUCACCAGCUUCAAGCUGGAGAGAAGCGAAUACUCCG_________ .................................................((((..((....))....)))).((((..(((((.....))))).))))...................... (-13.43 = -13.65 + 0.22)

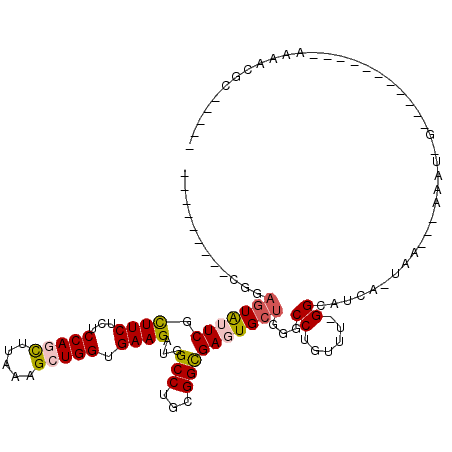
| Location | 11,491,008 – 11,491,102 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 67.26 |
| Mean single sequence MFE | -29.22 |
| Consensus MFE | -17.55 |
| Energy contribution | -17.75 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.09 |
| Structure conservation index | 0.60 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.529101 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11491008 94 - 23771897 ---------CGGAAUAUUCGCUUCUCUCCAGCUUAAAGCUGGUGAAGAUGGCCUGCGGCGAGUGCUGGGCCUAUUUUGGAAGCA-UAA---AAAUUGA--------A-AAAAAGCG---- ---------..........(((((((.(((((.....))))).)(((((((((..((((....)))))))).))))))))))).-...---.......--------.-........---- ( -29.00) >DroPse_CAF1 12291 93 - 1 CAGGCGCACUUGAGUGCUCAUUUCUUUCCAAUUUGAAGCUGGUGAAGAUGGCCUGGGGCGAAUGCUGAGCCUGCG--GGCAACA-GAA---AGA-----------------GGUCU---- .((((((((....))))....(((((((..........((.....))...(((((((((.........)))).))--)))....-)))---)))-----------------)))))---- ( -27.90) >DroSec_CAF1 6068 101 - 1 ---------CGGCGUAUUCGCUUCUCUCCAGCUUAAAGCUGGUGAAGAUGGCCUGCGGUGAGUGCUGGGCCUGUUUUGGGAUAAAUAAAAUAAAUCG---------A-AAAACGCGUACA ---------..((((.((((((((...(((((.....))))).))))..((((((((.....))).)))))(((((((.......)))))))...))---------)-)..))))..... ( -31.30) >DroPer_CAF1 2528 93 - 1 CAGGCGCACUUGAGUGCUCAUUUCUUUCCAAUUUAAAGCUGGUGAAGAUGGCCUGGGGCGAAUGCUGAGCCUGCU--GGCAACA-GAA---AGA-----------------GGUCU---- (((((((((....)))).....((((((((.........))).)))))..)))))((((.........)))).((--(....))-)..---...-----------------.....---- ( -28.80) >DroEre_CAF1 1732 95 - 1 ---------CGGAGUAUUCGCUUCGCUCCAGCUUGAAGCUGGUGAAGAUGGCCUGCGGCGAGUGCUGCGCCUGUAUGGGCAUCA-UAA---AAAUAGA----------AAAACGC--AUA ---------.(((((....)))))((.(((((.....)))))....((((.(((((((((.......)))).))).)).)))).-...---.......----------.....))--... ( -31.90) >DroYak_CAF1 2322 105 - 1 ---------CAGAGUAUUCGCUUCUCUCCAGCUUGAAGCUGGUGAAGAUGGCCUGCGGCGAGUGCUGGGCCUGUGGGGGGAUCG-UAA---AAAAAAAAAAAAUGGAAAAAACGU--AUA ---------..((...(((((.((((.(((((.....))))).).))).((((..((((....)))))))).)))))....)).-...---........................--... ( -26.40) >consensus _________CGGAGUAUUCGCUUCUCUCCAGCUUAAAGCUGGUGAAGAUGGCCUGCGGCGAGUGCUGGGCCUGUUU_GGCAUCA_UAA___AAAU_G___________AAAACGC_____ ............(((((((.((((...(((((.....))))).))))...(((...))))))))))...((......))......................................... (-17.55 = -17.75 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:10:51 2006