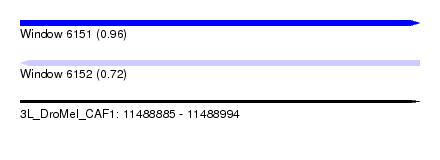
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 11,488,885 – 11,488,994 |
| Length | 109 |
| Max. P | 0.956676 |

| Location | 11,488,885 – 11,488,994 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.25 |
| Mean single sequence MFE | -32.86 |
| Consensus MFE | -22.86 |
| Energy contribution | -22.78 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.956676 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11488885 109 + 23771897 ACCUCCAUCGAUGGCUUCUUCAAUCGGAAACGGGGGCGUCCGCCAAAGAAUCGCUUUGUGGAGGUCUACAAGAGCGUGAGUAUUUUUCGA-----------AAUAUGUUAGGAUGCAUUU ..((((...(((..........)))(....)))))((((((..........((((((((((....)))).))))))...((((((....)-----------)))))....)))))).... ( -30.60) >DroPse_CAF1 10572 84 + 1 ACCUCCAUCGAUGGGUUCUUCAAUCGGAAACGGGGCCGUCCGCCCAAGAACCGUUUUGUGGAGGUAUACAAGAGCGUAAGUAGA------------------------------------ ((((((((.((..(((((((....((....))((((.....)))))))))))..)).))))))))........((....))...------------------------------------ ( -34.20) >DroSec_CAF1 3865 120 + 1 ACCUCCAUCGAUGGCUUCUUCAAUCGGAAACGGGGGCGUCCGCCAAAGAAUCGCUUUGUGGAGGUCUACAAGAGCGUGAGUAUUUUUCGAAAAUAUUUUGAAAUAUAUUAGGAUGUAUUU (((((((.(((.((((((((...(((....))).(((....))).)))))..)))))))))))))........((((.(((((.(((((((.....))))))).)))))...)))).... ( -34.00) >DroPer_CAF1 871 84 + 1 ACCUCCAUCGAUGGGUUCUUCAAUCGGAAACGGGGCCGUCCGCCCAAGAACCGUUUUGUGGAGGUCUACAAGAGCGUAAGUAGA------------------------------------ .(((((((.((..(((((((....((....))((((.....)))))))))))..)).)))))))(((((..........)))))------------------------------------ ( -34.00) >DroYak_CAF1 97 104 + 1 ACCUCCAUCGAUGGCUUCUUCAAUCGGAAACGGGGGCGGCCGCCAAAGAAUCGCUUUGUGGAGGUCUACAAGAGCGUGAGUAUACUUAGA-----------A-----UUGGGAUGCACUU .....((((...((((.((((...((....)))))).)))).((((.....((((((((((....)))).))))))((((....))))..-----------.-----))))))))..... ( -31.50) >consensus ACCUCCAUCGAUGGCUUCUUCAAUCGGAAACGGGGGCGUCCGCCAAAGAAUCGCUUUGUGGAGGUCUACAAGAGCGUGAGUAUA_UU_GA___________A_____UU_GGAUG_A_UU ((((((((.(((((.(((((...(((....))).(((....))).)))))))).)).)))))))).(((......))).......................................... (-22.86 = -22.78 + -0.08)



| Location | 11,488,885 – 11,488,994 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.25 |
| Mean single sequence MFE | -26.24 |
| Consensus MFE | -20.74 |
| Energy contribution | -20.70 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.09 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.722049 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11488885 109 - 23771897 AAAUGCAUCCUAACAUAUU-----------UCGAAAAAUACUCACGCUCUUGUAGACCUCCACAAAGCGAUUCUUUGGCGGACGCCCCCGUUUCCGAUUGAAGAAGCCAUCGAUGGAGGU ....((.(((.........-----------((((..........((((.((((.(....).)))))))).(((((..(((((.((....)).)))).)..)))))....)))).))).)) ( -25.10) >DroPse_CAF1 10572 84 - 1 ------------------------------------UCUACUUACGCUCUUGUAUACCUCCACAAAACGGUUCUUGGGCGGACGGCCCCGUUUCCGAUUGAAGAACCCAUCGAUGGAGGU ------------------------------------...................(((((((......(((((((.((((((((....))..)))).)).)))))))......))))))) ( -27.60) >DroSec_CAF1 3865 120 - 1 AAAUACAUCCUAAUAUAUUUCAAAAUAUUUUCGAAAAAUACUCACGCUCUUGUAGACCUCCACAAAGCGAUUCUUUGGCGGACGCCCCCGUUUCCGAUUGAAGAAGCCAUCGAUGGAGGU ....((.(((.(((((........))))).((((..........((((.((((.(....).)))))))).(((((..(((((.((....)).)))).)..)))))....)))).))).)) ( -25.60) >DroPer_CAF1 871 84 - 1 ------------------------------------UCUACUUACGCUCUUGUAGACCUCCACAAAACGGUUCUUGGGCGGACGGCCCCGUUUCCGAUUGAAGAACCCAUCGAUGGAGGU ------------------------------------(((((..........)))))((((((......(((((((.((((((((....))..)))).)).)))))))......)))))). ( -30.30) >DroYak_CAF1 97 104 - 1 AAGUGCAUCCCAA-----U-----------UCUAAGUAUACUCACGCUCUUGUAGACCUCCACAAAGCGAUUCUUUGGCGGCCGCCCCCGUUUCCGAUUGAAGAAGCCAUCGAUGGAGGU (((.((.......-----.-----------...............)).)))....(((((((.....((((((((..((((..((....))..))).)..))))....)))).))))))) ( -22.60) >consensus AA_U_CAUCC_AA_____U___________UC_AA_UAUACUCACGCUCUUGUAGACCUCCACAAAGCGAUUCUUUGGCGGACGCCCCCGUUUCCGAUUGAAGAAGCCAUCGAUGGAGGU .......................................................(((((((.....((((((((..(((((((....))..)))).)..))))....)))).))))))) (-20.74 = -20.70 + -0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:10:48 2006