
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 11,465,941 – 11,466,048 |
| Length | 107 |
| Max. P | 0.980924 |

| Location | 11,465,941 – 11,466,048 |
|---|---|
| Length | 107 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 81.90 |
| Mean single sequence MFE | -27.33 |
| Consensus MFE | -16.30 |
| Energy contribution | -17.97 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.41 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969157 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
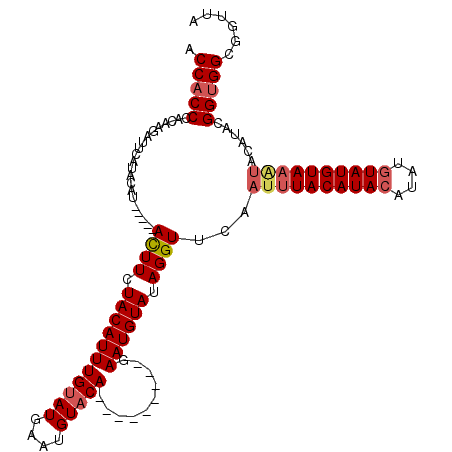
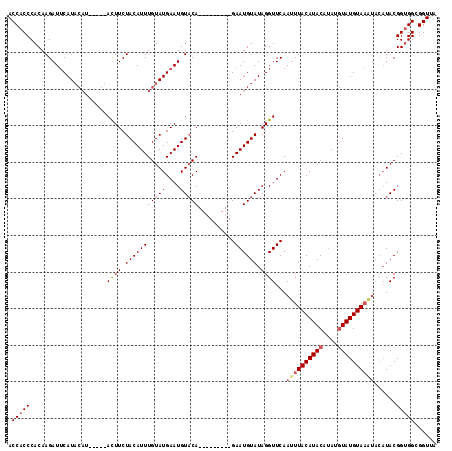
>3L_DroMel_CAF1 11465941 107 + 23771897 ACCACCCACAAGAUGCAU---------ACUUCUACAUUUGUAUGAAUGUUCAGAAUAUUCAGAAUGUAUAGGUUCAAUUUACAUACA-AUUUAUGUACAUACAUACGGCGGCGGUUA (((.(((.....(((...---------((((.(((((((...(((((((.....)))))))))))))).))))......((((((..-...))))))....)))...).)).))).. ( -20.10) >DroSec_CAF1 185480 103 + 1 ACCACCCACAAGAUUCAUACAU-----ACUUCUACAUUUGUAUGAAUGUACA---------GAAUGUAUAGGUUCAAUUUACAUACAUAUGUAUGUAACUACAUACGGUGGCGGUUA .(((((.....(((..((((((-----.((..(((((((....))))))).)---------).))))))..)))....((((((((....))))))))........)))))...... ( -27.80) >DroSim_CAF1 198750 104 + 1 ACCACCCACAAGAUUCAUACAUCUUGUAUUUCUACAUUUGUAUGAAUGUACA---------GAAUGUAUAGGUUCAAUUUACAUACAUAUGUAUGUAAG----UACGGUGGCGGUUA .(((((......(((((((((...((((....))))..)))))))))((((.---------((((......))))...((((((((....)))))))))----))))))))...... ( -34.10) >consensus ACCACCCACAAGAUUCAUACAU_____ACUUCUACAUUUGUAUGAAUGUACA_________GAAUGUAUAGGUUCAAUUUACAUACAUAUGUAUGUAAAUACAUACGGUGGCGGUUA .(((((.....................((((.(((((((((((....)))))..........)))))).))))...((((((((((....))))))))))......)))))...... (-16.30 = -17.97 + 1.67)

| Location | 11,465,941 – 11,466,048 |
|---|---|
| Length | 107 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 81.90 |
| Mean single sequence MFE | -28.53 |
| Consensus MFE | -17.65 |
| Energy contribution | -20.10 |
| Covariance contribution | 2.45 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.40 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.980924 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
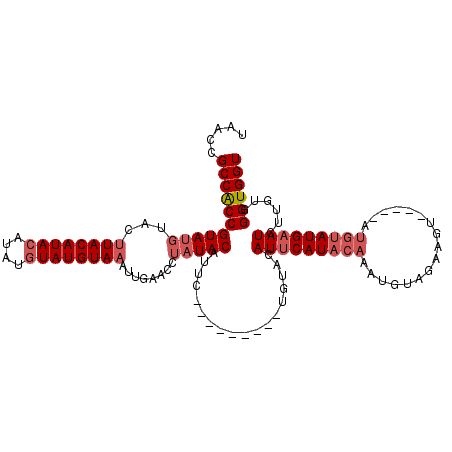
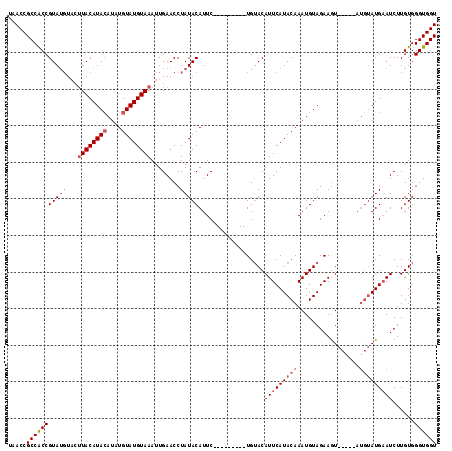
>3L_DroMel_CAF1 11465941 107 - 23771897 UAACCGCCGCCGUAUGUAUGUACAUAAAU-UGUAUGUAAAUUGAACCUAUACAUUCUGAAUAUUCUGAACAUUCAUACAAAUGUAGAAGU---------AUGCAUCUUGUGGGUGGU .....(((((((((((((((((((.....-)))))))).........)))))....(((((.........)))))((((((((((.....---------.))))).))))))))))) ( -23.10) >DroSec_CAF1 185480 103 - 1 UAACCGCCACCGUAUGUAGUUACAUACAUAUGUAUGUAAAUUGAACCUAUACAUUC---------UGUACAUUCAUACAAAUGUAGAAGU-----AUGUAUGAAUCUUGUGGGUGGU .....(((((((.(((((.((((((((....))))))))..........))))).)---------..((((((((((((.((......))-----.))))))))...)))))))))) ( -28.70) >DroSim_CAF1 198750 104 - 1 UAACCGCCACCGUA----CUUACAUACAUAUGUAUGUAAAUUGAACCUAUACAUUC---------UGUACAUUCAUACAAAUGUAGAAAUACAAGAUGUAUGAAUCUUGUGGGUGGU .....(((((((((----(((((((((....))))))))...(((........)))---------.))))(((((((((..((((....))))...)))))))))......)))))) ( -33.80) >consensus UAACCGCCACCGUAUGUACUUACAUACAUAUGUAUGUAAAUUGAACCUAUACAUUC_________UGUACAUUCAUACAAAUGUAGAAGU_____AUGUAUGAAUCUUGUGGGUGGU .....(((((((((((...((((((((....))))))))........)))))..................(((((((((.................)))))))))......)))))) (-17.65 = -20.10 + 2.45)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:10:44 2006