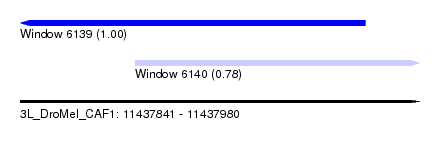
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 11,437,841 – 11,437,980 |
| Length | 139 |
| Max. P | 0.997679 |

| Location | 11,437,841 – 11,437,961 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.00 |
| Mean single sequence MFE | -39.22 |
| Consensus MFE | -35.80 |
| Energy contribution | -36.40 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.91 |
| SVM RNA-class probability | 0.997679 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11437841 120 - 23771897 CAGCUUCAACUGCAACUGCGUCAAGGGCAUUAAGCCCAACAAGGAGAAGCUGGCAAAGAUCGUGAAUGAGUCGCUAAUGCUGGUGACAGCCCUCAAUCCGCACAUUGGCUAUGACAAGUC (((((((..((((....)).....((((.....))))....))..)))))))((.......((((.....))))....((((....)))).........)).....((((......)))) ( -36.10) >DroSec_CAF1 157303 120 - 1 CAGCUUCAACUGCAACUGCGUCAAAGGCAUUAAGCCUAACAAGGAGAAGCUGGCCAAGAUCGUGAAUGAGUCGCUAAUGCUGGUGACAGCCCUCAAUCCGCACAUUGGCUAUGACAAGUC (((((((..((((....)).....((((.....))))....))..)))))))(((((...((.((.((((........((((....)))).)))).))))....)))))........... ( -38.90) >DroSim_CAF1 168931 120 - 1 CAGCUUCAAUUGCAACUGCGUCAAAGGCAUUAAACCCAACAAGGAGAAGCUGGCCAAGAUCGUGAAUGAGUCGCUAAUGCUGGUGACAGCACUCAAUCCGCACAUUGGCUAUGACAAGUC (((((((....((....))((.....)).......((.....)).)))))))(((((...((.((.(((((.......((((....))))))))).))))....)))))........... ( -36.01) >DroEre_CAF1 168527 120 - 1 CAGCUUCAACUGCAACUGCGUCAAGGGCAUUCGGCCCAACAAGGAGAAGCUGGCCAAGAUCGUGAACGAGUCGCUAAUGCUGGUGACAGCCCUCAAUCCGCACAUUGGCUAUGACAAGUC (((((((..((((....)).....((((.....))))....))..)))))))((((((((((....))).))......((((....))))..............)))))........... ( -40.90) >DroYak_CAF1 155692 120 - 1 CAGCUUCAACUGCAACUGCGUCAAGGGCAUCAAGCCCAACAAGGAGAAGCUGGCCAAGAUCGUGAACGAGUCGCUGAUGCUGGUGACAGCCCUCAAUCCGCACAUUGGCUAUGACAAGUC (((((((..((((....)).....((((.....))))....))..)))))))((((((((((....))).))((.(((((((....)))).....))).))...)))))........... ( -44.20) >consensus CAGCUUCAACUGCAACUGCGUCAAGGGCAUUAAGCCCAACAAGGAGAAGCUGGCCAAGAUCGUGAAUGAGUCGCUAAUGCUGGUGACAGCCCUCAAUCCGCACAUUGGCUAUGACAAGUC (((((((..((((....)).....((((.....))))....))..)))))))(((((...((.((.((((........((((....)))).)))).))))....)))))........... (-35.80 = -36.40 + 0.60)

| Location | 11,437,881 – 11,437,980 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 92.93 |
| Mean single sequence MFE | -29.52 |
| Consensus MFE | -27.10 |
| Energy contribution | -26.58 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.777010 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11437881 99 + 23771897 GCAUUAGCGACUCAUUCACGAUCUUUGCCAGCUUCUCCUUGUUGGGCUUAAUGCCCUUGACGCAGUUGCAGUUGAAGCUGAUGCACCCAUCAGCUAGGA ((((((((((.((......))))....(((((........))))))).))))))(((..((((....)).))..)((((((((....)))))))).)). ( -30.60) >DroSec_CAF1 157343 99 + 1 GCAUUAGCGACUCAUUCACGAUCUUGGCCAGCUUCUCCUUGUUAGGCUUAAUGCCUUUGACGCAGUUGCAGUUGAAGCUGAUGCACCCAUCAGCUAGAA ...(((((((.....))..(((....(((((((((..((.(((((((.....))))..)))((....))))..)))))))..))....))).))))).. ( -27.00) >DroSim_CAF1 168971 99 + 1 GCAUUAGCGACUCAUUCACGAUCUUGGCCAGCUUCUCCUUGUUGGGUUUAAUGCCUUUGACGCAGUUGCAAUUGAAGCUGAUGCACCCAUCAGCUAGAA ......(((((................(((((........)))))......(((.......))))))))......((((((((....)))))))).... ( -25.40) >DroEre_CAF1 168567 99 + 1 GCAUUAGCGACUCGUUCACGAUCUUGGCCAGCUUCUCCUUGUUGGGCCGAAUGCCCUUGACGCAGUUGCAGUUGAAGCUGAUGCAGCCAUCCGCUACGA ....(((((..(((....)))...(((((((((((..((.(((((((.....))))..)))((....))))..))))))).....))))..)))))... ( -31.60) >DroYak_CAF1 155732 99 + 1 GCAUCAGCGACUCGUUCACGAUCUUGGCCAGCUUCUCCUUGUUGGGCUUGAUGCCCUUGACGCAGUUGCAGUUGAAGCUGAUGCAACCAUCCGCUAUAA ((((((((((.(((....))))).....(((((.(..((((((((((.....))))..)))).))..).)))))..))))))))............... ( -33.00) >consensus GCAUUAGCGACUCAUUCACGAUCUUGGCCAGCUUCUCCUUGUUGGGCUUAAUGCCCUUGACGCAGUUGCAGUUGAAGCUGAUGCACCCAUCAGCUAGAA ((((((((((.((......)))).....(((((.(..((((((((((.....))))..)))).))..).)))))..))))))))............... (-27.10 = -26.58 + -0.52)

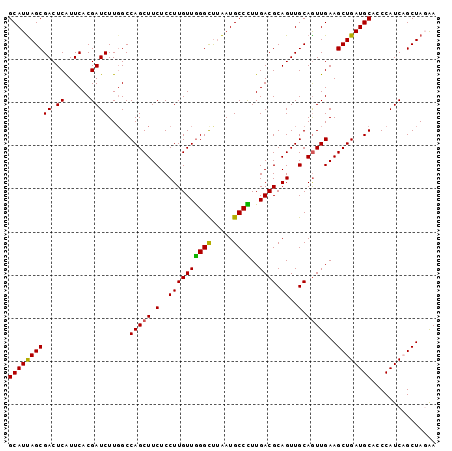
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:10:36 2006