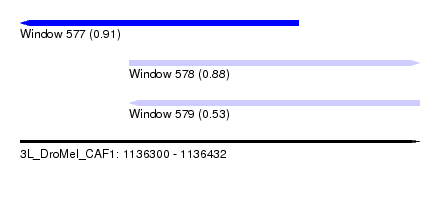
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,136,300 – 1,136,432 |
| Length | 132 |
| Max. P | 0.907707 |

| Location | 1,136,300 – 1,136,392 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 80.51 |
| Mean single sequence MFE | -21.57 |
| Consensus MFE | -13.44 |
| Energy contribution | -13.58 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.907707 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
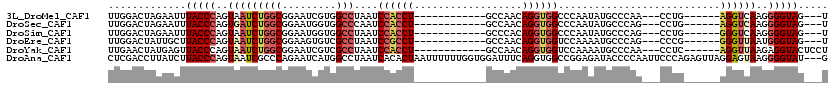
>3L_DroMel_CAF1 1136300 92 - 23771897 CUAAUCCACCUGCCAACAGGUGGCCCAAUAUGCCCAACCUGAGGUCAAGGGGUAG---UCGUAGAAG---CAAAUUACACAAAUUUAAAAAUAAUAAA (((..(((((((....))))))).......(((((.(((...)))....))))).---...)))...---............................ ( -19.10) >DroPse_CAF1 36120 81 - 1 AG-GUGCA-GAGCC--CACGGGGCCCAAUAUGCCCCA-------UCGAGGGGUGG---GCA---AAGGCACAAAUUACACAAAUUUAAAAAUAAUAAA ..-((((.-..(((--((((((((.......))))).-------.(....)))))---)).---...))))........................... ( -26.80) >DroSec_CAF1 29521 89 - 1 CCAAUCCACCUGCCAACAGGUGGCCCAAUAUGCCCAGCCUGAGGUCAAGGGGUAG---UCG---AAG---CAAAUUACACGAAUUUAAAAAUAAUAAA .....(((((((....))))))).......(((((.(((...)))....))))).---...---...---............................ ( -19.00) >DroSim_CAF1 27333 89 - 1 CUAAUCCACCUGCCCACAGGUGGCCCAAUAUGCCCAGCCUGGGGUCAAGGGGUAG---UCA---AAG---CAAAUUACACAAAUUUAAAAAUAAUAAA ........(((((((.((((((((.......)))..)))))))))..))).((((---(..---...---...))))).................... ( -22.10) >DroEre_CAF1 27493 89 - 1 CUAAUCCGCCUGCCAACAGGUGGUCCAAAAUGCCCAGCCCGGGGUUAAUGGGUAG---UCA---GAG---CAAAUUACACAGAUUUAAAAAUAAUAAA .....(((((((....))))))).(((....((((......))))...)))((((---(..---...---...))))).................... ( -21.20) >DroYak_CAF1 30395 92 - 1 CUAAUCCACCUGCCAACAGGUGGUCCAAAAUGCCCAACCUCAGGUUAAGAGGUACUCCUCA---GAG---CAAAUUACACGAAUUUAAUAAUAAUAAA .....(((((((....))))))).......(((.(((((...))))..((((....)))).---).)---)).......................... ( -21.20) >consensus CUAAUCCACCUGCCAACAGGUGGCCCAAUAUGCCCAACCUGAGGUCAAGGGGUAG___UCA___AAG___CAAAUUACACAAAUUUAAAAAUAAUAAA .....(((((((....))))))).......(((((.(((...)))....)))))............................................ (-13.44 = -13.58 + 0.14)

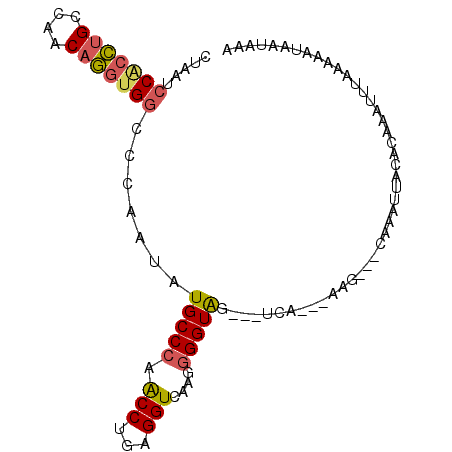
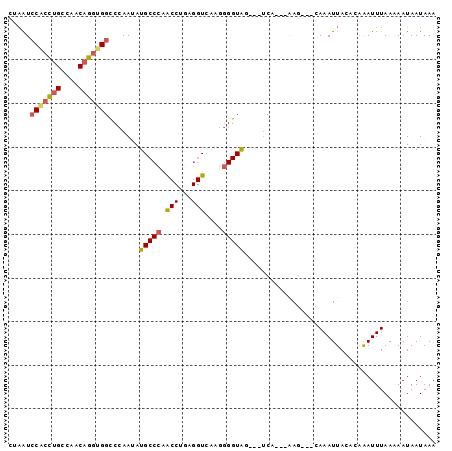
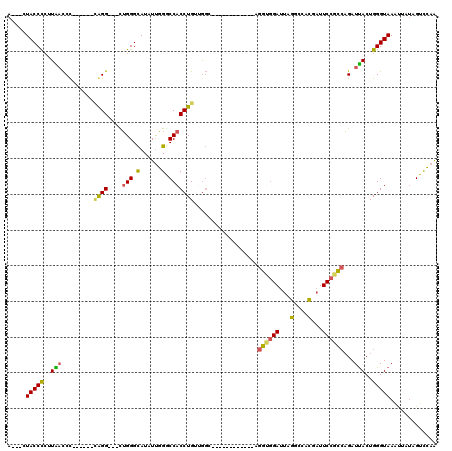
| Location | 1,136,336 – 1,136,432 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.23 |
| Mean single sequence MFE | -33.18 |
| Consensus MFE | -20.32 |
| Energy contribution | -19.80 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.878235 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
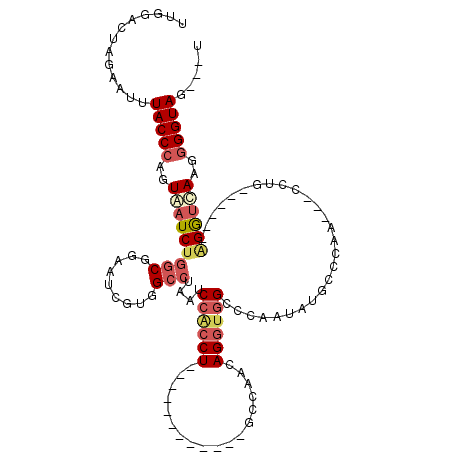
>3L_DroMel_CAF1 1136336 96 + 23771897 A---CUACCCCUUGACCU------CAGG---UUGGGCAUAUUGGGCCACCUGUUGGC------------AGGUGGAUUAGGCCACGAUUCCGCCAGAUUACUGGGUAAAUUCUAGUCCAA .---......((((.((.------((((---(.((.(......).)))))))..)))------------)))(((((((((...((....))((((....))))......))))))))). ( -31.30) >DroSec_CAF1 29554 96 + 1 A---CUACCCCUUGACCU------CAGG---CUGGGCAUAUUGGGCCACCUGUUGGC------------AGGUGGAUUGGGCCACCAUUCCGCCAGAUCACUGGGUAAAUUCUAGUCCAA .---.(((((..(((((.------((((---.(((.(......).)))))))..)).------------.((((((.(((....))).))))))...)))..)))))............. ( -37.10) >DroSim_CAF1 27366 96 + 1 A---CUACCCCUUGACCC------CAGG---CUGGGCAUAUUGGGCCACCUGUGGGC------------AGGUGGAUUAGGCCACCAUUCCGCCAGAUUACUGGGUAAAUUCUAGUCCAA .---.(((((..((.(((------((((---.(((.(......).))))))).))))------------)((((((...((...))..))))))........)))))............. ( -34.50) >DroEre_CAF1 27526 96 + 1 A---CUACCCAUUAACCC------CGGG---CUGGGCAUUUUGGACCACCUGUUGGC------------AGGCGGAUUAGGCGACACUUCCGCCAGAUUACUGGGUAAGCAAUAGUCCAA .---.((((((.(((((.------((((---.(((..........)))))))..)).------------.((((((...(....)...))))))...))).))))))............. ( -30.30) >DroYak_CAF1 30428 99 + 1 AGGAGUACCUCUUAACCU------GAGG---UUGGGCAUUUUGGACCACCUGUUGGC------------AGGUGGAUUAGGCGACGAUUCCGCCAGAUUACUGGGUAACUCAUAGUUCAA ..(((((((((((((((.------..))---))))).........(((((((....)------------))))))....((((.......))))........)))).))))......... ( -33.00) >DroAna_CAF1 32700 117 + 1 C---AUACCCCUUACUCCUAACUCUGGGAAUUGGGGUAUCUCCGGCCACCUGAAAUCCACCAAAAAAUUAGUGUGAUUAGGCCAUGAUUCUGGGCGAUUACUGGGUAAGAUAAGGUCGAG .---(((((((....(((((....)))))...)))))))...(((((.(((((..(((((..........))).)))))))((((((((......))))).))).........))))).. ( -32.90) >consensus A___CUACCCCUUAACCC______CAGG___CUGGGCAUAUUGGGCCACCUGUUGGC____________AGGUGGAUUAGGCCACGAUUCCGCCAGAUUACUGGGUAAAUUAUAGUCCAA .....(((((..(((.........((((....(((.(......).)))))))..................((((((...(....)...))))))...)))..)))))............. (-20.32 = -19.80 + -0.52)

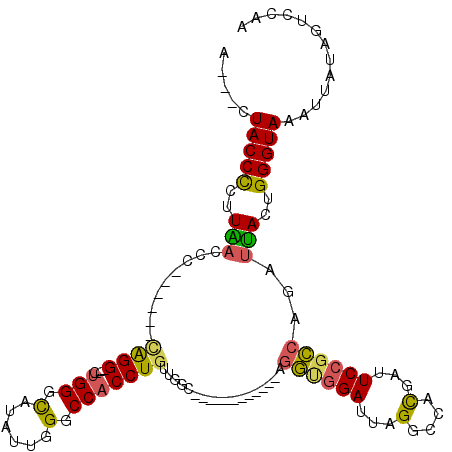
| Location | 1,136,336 – 1,136,432 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.23 |
| Mean single sequence MFE | -33.05 |
| Consensus MFE | -16.20 |
| Energy contribution | -17.29 |
| Covariance contribution | 1.09 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.49 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.527822 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
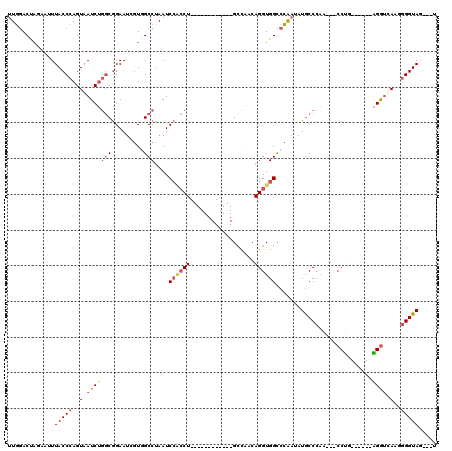
>3L_DroMel_CAF1 1136336 96 - 23771897 UUGGACUAGAAUUUACCCAGUAAUCUGGCGGAAUCGUGGCCUAAUCCACCU------------GCCAACAGGUGGCCCAAUAUGCCCAA---CCUG------AGGUCAAGGGGUAG---U ............((((((..(.(((((((((....((((......))))))------------)))..((((((((.......)))..)---))))------)))).)..))))))---. ( -31.10) >DroSec_CAF1 29554 96 - 1 UUGGACUAGAAUUUACCCAGUGAUCUGGCGGAAUGGUGGCCCAAUCCACCU------------GCCAACAGGUGGCCCAAUAUGCCCAG---CCUG------AGGUCAAGGGGUAG---U ............((((((..((((((((((....(((((......))))))------------)))..((((((((.......)))..)---))))------))))))..))))))---. ( -38.30) >DroSim_CAF1 27366 96 - 1 UUGGACUAGAAUUUACCCAGUAAUCUGGCGGAAUGGUGGCCUAAUCCACCU------------GCCCACAGGUGGCCCAAUAUGCCCAG---CCUG------GGGUCAAGGGGUAG---U ..((.(((..((((..((((....))))..))))..)))))....((.(((------------((((.((((((((.......)))..)---))))------))))..)))))...---. ( -33.10) >DroEre_CAF1 27526 96 - 1 UUGGACUAUUGCUUACCCAGUAAUCUGGCGGAAGUGUCGCCUAAUCCGCCU------------GCCAACAGGUGGUCCAAAAUGCCCAG---CCCG------GGGUUAAUGGGUAG---U ((((((.((((((.....))))))..(((((.....))))).....(((((------------(....))))))))))))..(((((((---((..------.)))...)))))).---. ( -33.00) >DroYak_CAF1 30428 99 - 1 UUGAACUAUGAGUUACCCAGUAAUCUGGCGGAAUCGUCGCCUAAUCCACCU------------GCCAACAGGUGGUCCAAAAUGCCCAA---CCUC------AGGUUAAGAGGUACUCCU .........((((.(((...(((((((((((.....)))))....((((((------------(....)))))))..............---....------))))))...))))))).. ( -29.80) >DroAna_CAF1 32700 117 - 1 CUCGACCUUAUCUUACCCAGUAAUCGCCCAGAAUCAUGGCCUAAUCACACUAAUUUUUUGGUGGAUUUCAGGUGGCCGGAGAUACCCCAAUUCCCAGAGUUAGGAGUAAGGGGUAU---G (((....................((.....))....(((((.((((.((((((....))))))))))......))))))))(((((((.(((((........)))))..)))))))---. ( -33.00) >consensus UUGGACUAGAAUUUACCCAGUAAUCUGGCGGAAUCGUGGCCUAAUCCACCU____________GCCAACAGGUGGCCCAAUAUGCCCAA___CCUG______AGGUCAAGGGGUAG___U .............(((((..(((((((((.........)))....((((((..................))))))...........................))))))..)))))..... (-16.20 = -17.29 + 1.09)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:40:49 2006