| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 11,421,257 – 11,421,347 |
| Length | 90 |
| Max. P | 0.639050 |

| Location | 11,421,257 – 11,421,347 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 93.00 |
| Mean single sequence MFE | -21.38 |
| Consensus MFE | -20.38 |
| Energy contribution | -20.62 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.639050 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11421257 90 + 23771897 GAUGCGAGUGCUAAAGUGUGAGUGCGGUAGCAAGAGCCAACAACCGCAUCGAGGCCAUGUUGCACUGCAACCAAAAUAAA-UACAAAUAGU ..(((.(((((....((.(..(((((((.((....)).....)))))))..).))......))))))))...........-.......... ( -22.00) >DroSec_CAF1 140971 90 + 1 GAUGCGAGUGCUAAAGUGUGAGUGCGGUAGCAAGAGCCAACAACCGCAUCGAGGCCAUGUUGCACUGAAAUAAAAAUAAA-UACAAAUAGU ......(((((....((.(..(((((((.((....)).....)))))))..).))......)))))..............-.......... ( -18.20) >DroEre_CAF1 148561 90 + 1 GAUGCGAGUGCUAAAGUGUGAGUGCGGUAGCAAGAGCCAACAACCGCAUCGAGGCCAUGUUGCACUGCAACAAAAAUAAA-AAGGAAUAGU ..(((.(((((....((.(..(((((((.((....)).....)))))))..).))......))))))))...........-.......... ( -22.00) >DroYak_CAF1 139410 91 + 1 GAUGCGAGUGCUAAAGUGUGAGUGCGGUAGCAAGAGCCAACAACCGCAUCGAGGCCAUGUUGCACUGCAACAAAAGCAAAUAAAUAAUAAA ((((((.(..((........))..)(((.......)))......))))))...((..((((((...))))))...)).............. ( -23.30) >consensus GAUGCGAGUGCUAAAGUGUGAGUGCGGUAGCAAGAGCCAACAACCGCAUCGAGGCCAUGUUGCACUGCAACAAAAAUAAA_AACAAAUAGU ..(((.(((((....((.(..(((((((.((....)).....)))))))..).))......))))))))...................... (-20.38 = -20.62 + 0.25)


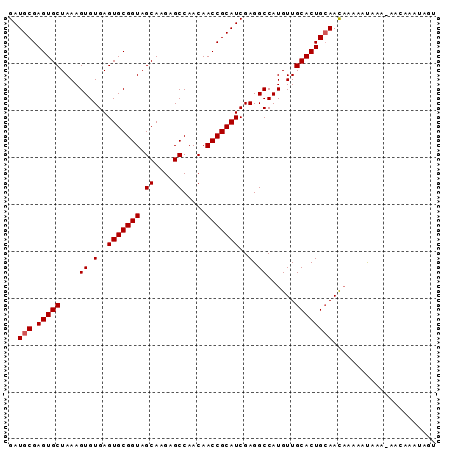
| Location | 11,421,257 – 11,421,347 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 93.00 |
| Mean single sequence MFE | -20.52 |
| Consensus MFE | -19.27 |
| Energy contribution | -19.52 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.608614 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11421257 90 - 23771897 ACUAUUUGUA-UUUAUUUUGGUUGCAGUGCAACAUGGCCUCGAUGCGGUUGUUGGCUCUUGCUACCGCACUCACACUUUAGCACUCGCAUC ..........-...........((((((((.....(....)((((((((....(((....))))))))).))........))))).))).. ( -20.90) >DroSec_CAF1 140971 90 - 1 ACUAUUUGUA-UUUAUUUUUAUUUCAGUGCAACAUGGCCUCGAUGCGGUUGUUGGCUCUUGCUACCGCACUCACACUUUAGCACUCGCAUC .(((((((((-((.(........).))))))).))))....((((((((....(((....))))))))).))........((....))... ( -18.10) >DroEre_CAF1 148561 90 - 1 ACUAUUCCUU-UUUAUUUUUGUUGCAGUGCAACAUGGCCUCGAUGCGGUUGUUGGCUCUUGCUACCGCACUCACACUUUAGCACUCGCAUC ..........-...........((((((((.....(....)((((((((....(((....))))))))).))........))))).))).. ( -20.90) >DroYak_CAF1 139410 91 - 1 UUUAUUAUUUAUUUGCUUUUGUUGCAGUGCAACAUGGCCUCGAUGCGGUUGUUGGCUCUUGCUACCGCACUCACACUUUAGCACUCGCAUC ..............(((..((((((...)))))).)))...((((((((....(((....))))))))).))........((....))... ( -22.20) >consensus ACUAUUUGUA_UUUAUUUUUGUUGCAGUGCAACAUGGCCUCGAUGCGGUUGUUGGCUCUUGCUACCGCACUCACACUUUAGCACUCGCAUC ......................((((((((.....(....)((((((((....(((....))))))))).))........))))).))).. (-19.27 = -19.52 + 0.25)

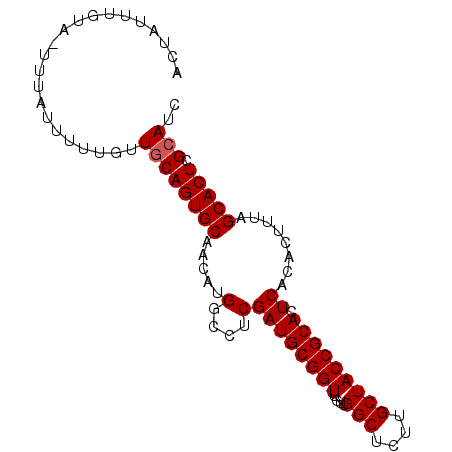
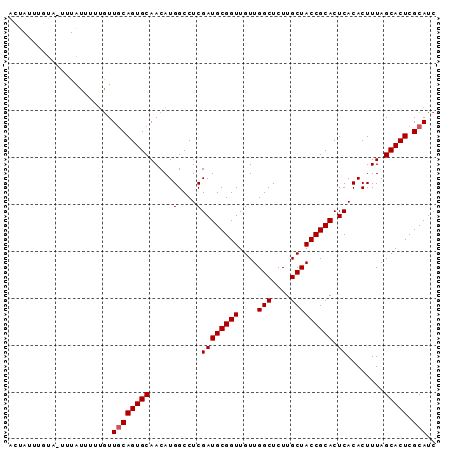
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:10:30 2006