| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 11,420,751 – 11,420,884 |
| Length | 133 |
| Max. P | 0.999479 |

| Location | 11,420,751 – 11,420,844 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 98.07 |
| Mean single sequence MFE | -21.28 |
| Consensus MFE | -20.18 |
| Energy contribution | -20.02 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.95 |
| SVM decision value | 3.64 |
| SVM RNA-class probability | 0.999479 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11420751 93 + 23771897 AACAA-AAACUGGGAGAAAAAUGCAAAGCAAACAAGCACCAAAUUGCAUUGCUGUAAAGCAUGUUUUGUGUUUGCUUUUUUGUAGAAUUUUUAA .....-...(((.(((((((((((((((((.....(((......)))..((((....)))))))))))))))...)))))).)))......... ( -21.40) >DroSec_CAF1 140476 93 + 1 AACAA-AAACUGGGAGAAAAAUGCAAAGCAAACAAGCACCAAAUUGCAUUGCUGUAAAGCAUGUUUUGUGUUUGCUUUUUUGUAGAAUUUUUAA .....-...(((.(((((((((((((((((.....(((......)))..((((....)))))))))))))))...)))))).)))......... ( -21.40) >DroSim_CAF1 151958 93 + 1 AACAA-AAACCGGGAGAAAAAUGCAAAGCAAACAAGCACCAAAUUGCAUUGCUGUAAAGCAUGUUUUGUGUUUGCUUUUUUGUAGAAUUUUUAA .((((-(((...((....((((((((((((.....(((......)))..((((....)))))))))))))))).)))))))))........... ( -20.90) >DroEre_CAF1 148051 92 + 1 AACAA-AAACUGGGGGAAAAAUGCAAAGCAAACAAGCACCAAAUUGCAUUGCUGUAAAGCAUGUUUUGUGUUUGCUU-UUUGUAGAAUUUUUAA .....-...(((.(((((((((((((((((.....(((......)))..((((....))))))))))))))))..))-))).)))......... ( -20.90) >DroYak_CAF1 138900 93 + 1 AACAAAAAACUGGGAGAAAAAUGCAAAGCAAACAAGCACCAAAUUGCAUUGCUGUAAAGCAUGUUUUGUGUUUGCUU-UUUGUAGAAUUUUUAA .........(((.(((((((((((((((((.....(((......)))..((((....))))))))))))))))..))-))).)))......... ( -21.80) >consensus AACAA_AAACUGGGAGAAAAAUGCAAAGCAAACAAGCACCAAAUUGCAUUGCUGUAAAGCAUGUUUUGUGUUUGCUUUUUUGUAGAAUUUUUAA .............(((((...(((((((((((((.(((......)))..((((....)))).......))))))))...)))))...))))).. (-20.18 = -20.02 + -0.16)

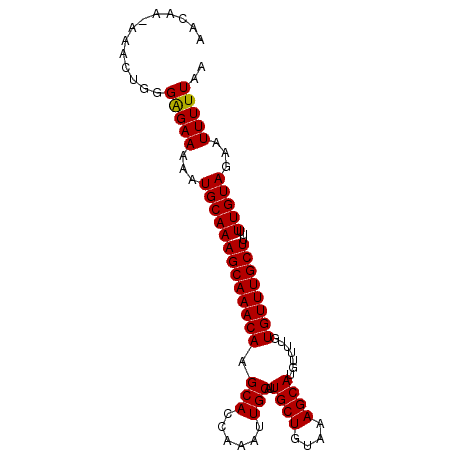

| Location | 11,420,751 – 11,420,844 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 98.07 |
| Mean single sequence MFE | -16.00 |
| Consensus MFE | -14.74 |
| Energy contribution | -15.14 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.836596 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11420751 93 - 23771897 UUAAAAAUUCUACAAAAAAGCAAACACAAAACAUGCUUUACAGCAAUGCAAUUUGGUGCUUGUUUGCUUUGCAUUUUUCUCCCAGUUU-UUGUU ................((((((((((.......((((....))))..(((......))).))))))))))..................-..... ( -15.90) >DroSec_CAF1 140476 93 - 1 UUAAAAAUUCUACAAAAAAGCAAACACAAAACAUGCUUUACAGCAAUGCAAUUUGGUGCUUGUUUGCUUUGCAUUUUUCUCCCAGUUU-UUGUU ................((((((((((.......((((....))))..(((......))).))))))))))..................-..... ( -15.90) >DroSim_CAF1 151958 93 - 1 UUAAAAAUUCUACAAAAAAGCAAACACAAAACAUGCUUUACAGCAAUGCAAUUUGGUGCUUGUUUGCUUUGCAUUUUUCUCCCGGUUU-UUGUU ................((((((((((.......((((....))))..(((......))).))))))))))..................-..... ( -15.90) >DroEre_CAF1 148051 92 - 1 UUAAAAAUUCUACAAA-AAGCAAACACAAAACAUGCUUUACAGCAAUGCAAUUUGGUGCUUGUUUGCUUUGCAUUUUUCCCCCAGUUU-UUGUU ...............(-(((((((((.......((((....))))..(((......))).))))))))))..................-..... ( -15.90) >DroYak_CAF1 138900 93 - 1 UUAAAAAUUCUACAAA-AAGCAAACACAAAACAUGCUUUACAGCAAUGCAAUUUGGUGCUUGUUUGCUUUGCAUUUUUCUCCCAGUUUUUUGUU ...........(((((-((((......((((.((((.....(((((.(((......)))....)))))..))))))))......))))))))). ( -16.40) >consensus UUAAAAAUUCUACAAAAAAGCAAACACAAAACAUGCUUUACAGCAAUGCAAUUUGGUGCUUGUUUGCUUUGCAUUUUUCUCCCAGUUU_UUGUU ................((((((((((.......((((....))))..(((......))).))))))))))........................ (-14.74 = -15.14 + 0.40)


| Location | 11,420,774 – 11,420,884 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 99.09 |
| Mean single sequence MFE | -20.20 |
| Consensus MFE | -18.92 |
| Energy contribution | -19.32 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.597474 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11420774 110 - 23771897 ACGGCCUAGAAGUUUUGCUCGCCAAACAGAAAAAAAUUGUUUAAAAAUUCUACAAAAAAGCAAACACAAAACAUGCUUUACAGCAAUGCAAUUUGGUGCUUGUUUGCUUU ..(((..((........)).)))((((((.......))))))..............((((((((((.......((((....))))..(((......))).)))))))))) ( -19.80) >DroSec_CAF1 140499 109 - 1 ACGGCCUAGAAGUUUUGCUCGCCAAACAGAAA-AAAUUGUUUAAAAAUUCUACAAAAAAGCAAACACAAAACAUGCUUUACAGCAAUGCAAUUUGGUGCUUGUUUGCUUU ..(((..((........)).)))((((((...-...))))))..............((((((((((.......((((....))))..(((......))).)))))))))) ( -20.30) >DroSim_CAF1 151981 109 - 1 ACGGCCUAGAAGUUUUGCUCGCCAAACAGAAA-AAAUUGUUUAAAAAUUCUACAAAAAAGCAAACACAAAACAUGCUUUACAGCAAUGCAAUUUGGUGCUUGUUUGCUUU ..(((..((........)).)))((((((...-...))))))..............((((((((((.......((((....))))..(((......))).)))))))))) ( -20.30) >DroEre_CAF1 148074 108 - 1 ACGGCCUAGAAGUUUUGCUCGCCAAACAGAAA-AAAUUGUUUAAAAAUUCUACAAA-AAGCAAACACAAAACAUGCUUUACAGCAAUGCAAUUUGGUGCUUGUUUGCUUU ..(((..((........)).)))((((((...-...)))))).............(-(((((((((.......((((....))))..(((......))).)))))))))) ( -20.30) >DroYak_CAF1 138924 108 - 1 ACGGCCUAGAAGUUUUGCUCGCCAAACAGAAA-AAAUUGUUUAAAAAUUCUACAAA-AAGCAAACACAAAACAUGCUUUACAGCAAUGCAAUUUGGUGCUUGUUUGCUUU ..(((..((........)).)))((((((...-...)))))).............(-(((((((((.......((((....))))..(((......))).)))))))))) ( -20.30) >consensus ACGGCCUAGAAGUUUUGCUCGCCAAACAGAAA_AAAUUGUUUAAAAAUUCUACAAAAAAGCAAACACAAAACAUGCUUUACAGCAAUGCAAUUUGGUGCUUGUUUGCUUU ..(((..((........)).)))((((((.......))))))..............((((((((((.......((((....))))..(((......))).)))))))))) (-18.92 = -19.32 + 0.40)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:10:29 2006