
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 11,406,574 – 11,406,750 |
| Length | 176 |
| Max. P | 0.743339 |

| Location | 11,406,574 – 11,406,684 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 87.37 |
| Mean single sequence MFE | -34.04 |
| Consensus MFE | -28.08 |
| Energy contribution | -28.84 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.644784 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11406574 110 - 23771897 CGUUGCUCUGAUGUCGCCAUUGAAUGAACAU--------AUGAAUAUUAAGAAACCGUGACGUAGAACGGAAAAUCGGACGACGACACGUGCGGUUCAUAGGACGGCGGCGACGUCAA ........((((((((((.(((.((((((..--------...............((((........)))).....((.(((......))).))))))))....))).)))))))))). ( -34.40) >DroSec_CAF1 126695 110 - 1 CGUUGCUCCGAUGUCGCCAUUGAAAGAACCU--------GGGAAUAUUAAGAAACCGUGACGUAGAACGGAAAAUCGGACGACGACACGUGCGGUUCAUAGUUCGGCGGCGAUGUCAA .........(((((((((.(((((.(((((.--------...............((((........))))......(.(((......))).))))))....))))).))))))))).. ( -33.90) >DroSim_CAF1 139308 110 - 1 CGUUGCUCCGAUGUCGCCAUUGAAAGAACCC--------AGGAAUAUUAAGAAACCGUGACGUAGAACGGAAAAUCGGACGACGACACGUGCGGUUCAUAGUUCGGCGGCGAUGUCAA .........(((((((((.(((((.((((((--------(.(............((((........))))....(((.....)))..).)).)))))....))))).))))))))).. ( -35.20) >DroEre_CAF1 133833 110 - 1 CGUUGCUCCGAUGUCGCCAUUGAAAAGGCGU--------AGGAAUAUUAAAAACCCGUGACGUAGAACGGAAAAUCGGACCUCGACACGUGCGGUUCAUAGGUCGGCGGCGACGUCAA ((((((((((((..((((........)))).--------...............((((........))))......(((((.((.....)).)))))....))))).))))))).... ( -33.40) >DroYak_CAF1 124609 110 - 1 CGUU--------GUCGCCAUUGAAAAGGCAUAAGAAUAUAGGAAUAUUAAGAAACCGUGACGUAGAACGGAAAAUCGGACGUCGACACGUGCGGUUCAUAGGUCGGCGGCGACGUCAA ((((--------((((((..(....)(((.((.((((.................((((........)))).....((.((((....)))).)))))).)).))))))))))))).... ( -33.30) >consensus CGUUGCUCCGAUGUCGCCAUUGAAAGAACAU________AGGAAUAUUAAGAAACCGUGACGUAGAACGGAAAAUCGGACGACGACACGUGCGGUUCAUAGGUCGGCGGCGACGUCAA .........(((((((((.((((...............................((((........))))..(((((.(((......))).)))))......)))).))))))))).. (-28.08 = -28.84 + 0.76)


| Location | 11,406,614 – 11,406,719 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 86.76 |
| Mean single sequence MFE | -27.43 |
| Consensus MFE | -19.66 |
| Energy contribution | -21.18 |
| Covariance contribution | 1.52 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.743339 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11406614 105 + 23771897 CCGAUUUUCCGUUCUACGUCACGGUUUCUUAAUAUUCAU--------AUGUUCAUUCAAUGGCGACAUCAGAGCAACGUGUCCCAUUGCAAUAUGAUAUGGCCUACGUGACGU ...............((((((((........((((.(((--------((.......((((((.(((((.........)))))))))))..)))))))))......)))))))) ( -27.24) >DroSec_CAF1 126735 105 + 1 CCGAUUUUCCGUUCUACGUCACGGUUUCUUAAUAUUCCC--------AGGUUCUUUCAAUGGCGACAUCGGAGCAACGUGUCCCAUUGCCAUAUGAUAUGGCCUACGUGACGU ...............((((((((................--------..........(((((.((((.((......)))))))))))((((((...))))))...)))))))) ( -30.50) >DroSim_CAF1 139348 105 + 1 CCGAUUUUCCGUUCUACGUCACGGUUUCUUAAUAUUCCU--------GGGUUCUUUCAAUGGCGACAUCGGAGCAACGUGUCCCAUUGCCAUAUGAUAUGGCCUACGUGACGU ...............((((((((................--------..........(((((.((((.((......)))))))))))((((((...))))))...)))))))) ( -30.50) >DroEre_CAF1 133873 102 + 1 CCGAUUUUCCGUUCUACGUCACGGGUUUUUAAUAUUCCU--------ACGCCUUUUCAAUGGCGACAUCGGAGCAACGUGUCCCAUUGCAAUAUGAU---GCCUACGUGACUU .................(((((((((.....(((((...--------.........((((((.((((.((......)))))))))))).)))))...---)))..)))))).. ( -24.32) >DroYak_CAF1 124649 103 + 1 CCGAUUUUCCGUUCUACGUCACGGUUUCUUAAUAUUCCUAUAUUCUUAUGCCUUUUCAAUGGCGAC--------AACGUGUCCCAUUGCCAUAUGAUA--GCCUACGUGACGU ...............((((((((((.....(((((....))))).....)))....((((((.(((--------.....)))))))))..........--......))))))) ( -24.60) >consensus CCGAUUUUCCGUUCUACGUCACGGUUUCUUAAUAUUCCU________AGGUUCUUUCAAUGGCGACAUCGGAGCAACGUGUCCCAUUGCCAUAUGAUAUGGCCUACGUGACGU ...............((((((((.............((((......))))......((((((.((((.((......)))))))))))).................)))))))) (-19.66 = -21.18 + 1.52)



| Location | 11,406,614 – 11,406,719 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 86.76 |
| Mean single sequence MFE | -28.60 |
| Consensus MFE | -21.86 |
| Energy contribution | -23.18 |
| Covariance contribution | 1.32 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.730984 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11406614 105 - 23771897 ACGUCACGUAGGCCAUAUCAUAUUGCAAUGGGACACGUUGCUCUGAUGUCGCCAUUGAAUGAACAU--------AUGAAUAUUAAGAAACCGUGACGUAGAACGGAAAAUCGG ((((((((..(((.((((((....((((((.....))))))..)))))).)))..((......)).--------................))))))))............... ( -29.00) >DroSec_CAF1 126735 105 - 1 ACGUCACGUAGGCCAUAUCAUAUGGCAAUGGGACACGUUGCUCCGAUGUCGCCAUUGAAAGAACCU--------GGGAAUAUUAAGAAACCGUGACGUAGAACGGAAAAUCGG ((((((((...((((((...))))))(((((((((((......)).)))).)))))..........--------................))))))))............... ( -29.60) >DroSim_CAF1 139348 105 - 1 ACGUCACGUAGGCCAUAUCAUAUGGCAAUGGGACACGUUGCUCCGAUGUCGCCAUUGAAAGAACCC--------AGGAAUAUUAAGAAACCGUGACGUAGAACGGAAAAUCGG ((((((((...((((((...))))))(((((((((((......)).)))).)))))..........--------................))))))))............... ( -29.60) >DroEre_CAF1 133873 102 - 1 AAGUCACGUAGGC---AUCAUAUUGCAAUGGGACACGUUGCUCCGAUGUCGCCAUUGAAAAGGCGU--------AGGAAUAUUAAAAACCCGUGACGUAGAACGGAAAAUCGG ..((((((..(((---(((.....((((((.....))))))...))))))(((........)))..--------................))))))................. ( -29.00) >DroYak_CAF1 124649 103 - 1 ACGUCACGUAGGC--UAUCAUAUGGCAAUGGGACACGUU--------GUCGCCAUUGAAAAGGCAUAAGAAUAUAGGAAUAUUAAGAAACCGUGACGUAGAACGGAAAAUCGG ((((((((....(--(.......(((((((.....))))--------)))(((........)))....................))....))))))))............... ( -25.80) >consensus ACGUCACGUAGGCCAUAUCAUAUGGCAAUGGGACACGUUGCUCCGAUGUCGCCAUUGAAAGAACAU________AGGAAUAUUAAGAAACCGUGACGUAGAACGGAAAAUCGG ((((((((..(((.(((((.....((((((.....))))))...))))).))).((....))............................))))))))............... (-21.86 = -23.18 + 1.32)

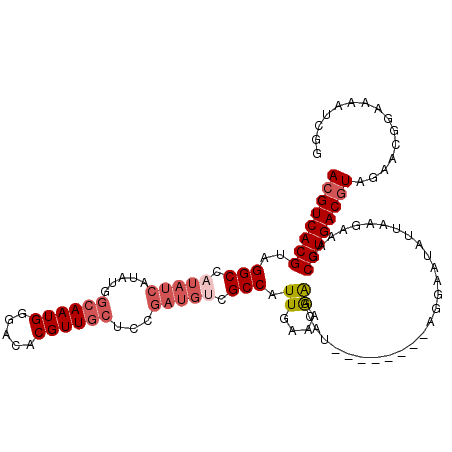

| Location | 11,406,653 – 11,406,750 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 70.47 |
| Mean single sequence MFE | -27.55 |
| Consensus MFE | -15.23 |
| Energy contribution | -15.98 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.599802 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11406653 97 + 23771897 -------AUGUUCAUUCAAUGGCGACAUC--AGAGCAACGUGUCCCAUUGCAAUAUGA---UA--UGGCCUACGUGACGUCUGCUACAACGUGCGGCGAUCAAUUGACCAA--------- -------....((((.((((((.(((((.--........)))))))))))....))))---..--(((.(....(((((.((((........)))))).)))...).))).--------- ( -25.60) >DroSec_CAF1 126774 97 + 1 -------AGGUUCUUUCAAUGGCGACAUC--GGAGCAACGUGUCCCAUUGCCAUAUGA---UA--UGGCCUACGUGACGUCUGCUACUACGUGUGGCGAUCAAUUGACCAA--------- -------.((((...(((((((.((((.(--(......)))))))))))((((((((.---..--((((..(((...)))..))))...))))))))))......))))..--------- ( -32.40) >DroSim_CAF1 139387 97 + 1 -------GGGUUCUUUCAAUGGCGACAUC--GGAGCAACGUGUCCCAUUGCCAUAUGA---UA--UGGCCUACGUGACGUCUGCUACUACGUGUGGCGAUCAAUUGACCAA--------- -------.((((...(((((((.((((.(--(......)))))))))))((((((((.---..--((((..(((...)))..))))...))))))))))......))))..--------- ( -32.20) >DroEre_CAF1 133912 94 + 1 -------ACGCCUUUUCAAUGGCGACAUC--GGAGCAACGUGUCCCAUUGCAAUAUGA---U-----GCCUACGUGACUUCUGCUGCUACGUGUGGCGAUCAAUUGACCAA--------- -------.........((((((.((((.(--(......))))))))))))((((.(((---(-----(((((((((.(.......).)))))).))).)))))))).....--------- ( -29.20) >DroYak_CAF1 124689 94 + 1 UAUUCUUAUGCCUUUUCAAUGGCGAC----------AACGUGUCCCAUUGCCAUAUGA---UA----GCCUACGUGACGUCUGCUGCAACGUGCGGCGAUCAAUUGACCAA--------- ................((((((.(((----------.....))))))))).((..(((---(.----((((((((..((.....))..))))).))).))))..)).....--------- ( -23.70) >DroAna_CAF1 117983 104 + 1 ----------------AAAUGGUGACGACAUAAAGGAACGUGCUCUUUUUUAAAAAAAGAAUACCCGACCUACGUGGCCUUCAUAUCCACGUGUGACUUUGCCUUGGUCAGAAUUACUUU ----------------....(....)(((...((((...(((.((((((.....)))))).)))....(.(((((((.........))))))).)......)))).)))........... ( -22.20) >consensus _______AGGUUCUUUCAAUGGCGACAUC__GGAGCAACGUGUCCCAUUGCAAUAUGA___UA__UGGCCUACGUGACGUCUGCUACUACGUGUGGCGAUCAAUUGACCAA_________ ................((((((.(((((...........))))))))))).................(((((((((...........)))))).)))....................... (-15.23 = -15.98 + 0.75)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:10:21 2006