| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 11,403,683 – 11,403,809 |
| Length | 126 |
| Max. P | 0.995839 |

| Location | 11,403,683 – 11,403,786 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 85.84 |
| Mean single sequence MFE | -24.20 |
| Consensus MFE | -18.66 |
| Energy contribution | -18.26 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.950000 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
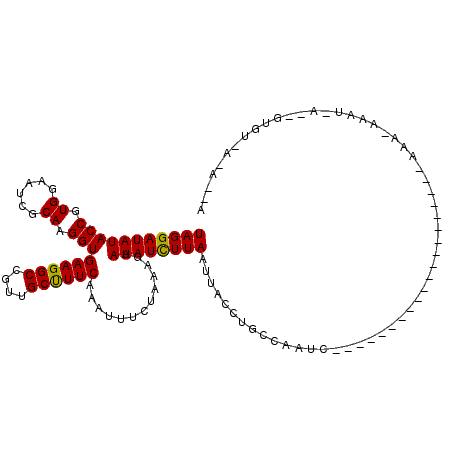
>3L_DroMel_CAF1 11403683 103 + 23771897 CCCGAAUGCGAUCCUAUAAUAUCAAUUAGGAUAUACCGUGGAAACACAAGGUGAAGGCCAUUGCUUUCACAUUUCUAAAGAAUAUCUUAAUUACCUGCCAAUG--- .......(((((........)))(((((((((((...(((....)))...((((((((....))))))))...........)))))))))))....)).....--- ( -27.10) >DroSec_CAF1 123914 101 + 1 CC--AUUGCGAUCCUAUAAUAUCAAUUAGGAUAUACCGUGGAAUCGCAAGGUGAAGGCCGUUGCUUUCACAUUUCUAAAGAAUAUCUUAAUUAGCUGCCAAUC--- ..--...(((((........)))(((((((((((.(..(((((.(....)((((((((....))))))))..)))))..).))))))))))).))........--- ( -27.00) >DroSim_CAF1 136040 101 + 1 CC--AAUGCGAUCCUACAAUAUCAAUUAGGAUAUACCGUGGAAUCGCAAGGUGAAGGCCGUUGCUUUCAAAUUUCUAAAGAAUAUCUUAAUUAGCUGCCAAUC--- ..--..((((((.((((.(((((......)))))...)))).)))))).(((((((((....)))))).........(((.....)))........)))....--- ( -24.50) >DroEre_CAF1 130890 97 + 1 CCA---UGAGAUCCCAUAAUAUCAAUUAGGAUAUACCGUGGAAUAACAAGGUGAAGGCCGUUGCUUUCAAAUUCUUAAGGCAUAUUUUAAUUACCUGCUG------ ...---.................(((((((((((.((..(((((.......(((((((....))))))).)))))...)).)))))))))))........------ ( -19.20) >DroYak_CAF1 121644 106 + 1 CCAAAAUGAGAUCCUAUAAUAUCAAUUAGGAUAUACCGUGGAAAAGCAAGGUGAAGGCCGUUGCCUUCAAAAUCUUAAAGCAUAUUUUAAUUACCUGCCAAUAUCU (((.......((((((..........))))))......)))....(((.(((((((((....))))))......((((((....))))))..))))))........ ( -23.22) >consensus CC__AAUGCGAUCCUAUAAUAUCAAUUAGGAUAUACCGUGGAAUCGCAAGGUGAAGGCCGUUGCUUUCAAAUUUCUAAAGAAUAUCUUAAUUACCUGCCAAUC___ .......................((((((((((((((.((......)).)))((((((....)))))).............))))))))))).............. (-18.66 = -18.26 + -0.40)


| Location | 11,403,709 – 11,403,809 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 68.35 |
| Mean single sequence MFE | -23.00 |
| Consensus MFE | -15.56 |
| Energy contribution | -15.16 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.68 |
| SVM decision value | 2.62 |
| SVM RNA-class probability | 0.995839 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11403709 100 + 23771897 UAGGAUAUACCGUGGAAACACAAGGUGAAGGCCAUUGCUUUCACAUUUCUAAAGAAUAUCUUAAUUACCUGCCAAUG--------------------CAAGAAAUUAGUGUGUGAAAGCA ..((...(((((((....)))..))))....))..(((((((((((..((((((.....))).....(.(((....)--------------------)).)....))).))))))))))) ( -28.50) >DroSec_CAF1 123938 79 + 1 UAGGAUAUACCGUGGAAUCGCAAGGUGAAGGCCGUUGCUUUCACAUUUCUAAAGAAUAUCUUAAUUAGCUGCCAAUC--------------------UA--------------------- ((((((((.(..(((((.(....)((((((((....))))))))..)))))..).))))))))..............--------------------..--------------------- ( -22.10) >DroSim_CAF1 136064 79 + 1 UAGGAUAUACCGUGGAAUCGCAAGGUGAAGGCCGUUGCUUUCAAAUUUCUAAAGAAUAUCUUAAUUAGCUGCCAAUC--------------------UA--------------------- ((((((((.(..(((((((....))(((((((....)))))))...)))))..).))))))))..............--------------------..--------------------- ( -19.20) >DroEre_CAF1 130913 100 + 1 UAGGAUAUACCGUGGAAUAACAAGGUGAAGGCCGUUGCUUUCAAAUUCUUAAGGCAUAUUUUAAUUACCUGCUG--------------------AUUAUAUAAAUGACAGUGUAAAAAAA ((((((((.((..(((((.......(((((((....))))))).)))))...)).)))))))).((((..((((--------------------.((((....))))))))))))..... ( -21.20) >DroYak_CAF1 121670 120 + 1 UAGGAUAUACCGUGGAAAAGCAAGGUGAAGGCCGUUGCCUUCAAAAUCUUAAAGCAUAUUUUAAUUACCUGCCAAUAUCUCAUAUAACUACUGUAUUAUACAAAUGAGAGUGUGAGAACA ((((((..(((.((......)).)))((((((....))))))...))))))..(((.............))).....((((((((..((..((((...))))....)).))))))))... ( -24.02) >consensus UAGGAUAUACCGUGGAAUCGCAAGGUGAAGGCCGUUGCUUUCAAAUUUCUAAAGAAUAUCUUAAUUACCUGCCAAUC____________________AAA_AAAU_A__GUGU_A_A__A (((((((((((.((......)).)))((((((....)))))).............))))))))......................................................... (-15.56 = -15.16 + -0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:10:16 2006