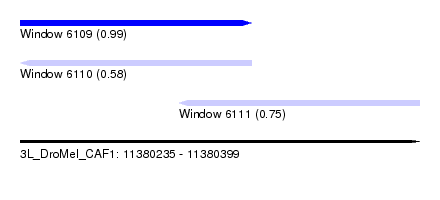
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 11,380,235 – 11,380,399 |
| Length | 164 |
| Max. P | 0.987755 |

| Location | 11,380,235 – 11,380,330 |
|---|---|
| Length | 95 |
| Sequences | 3 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 97.19 |
| Mean single sequence MFE | -29.93 |
| Consensus MFE | -29.00 |
| Energy contribution | -29.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.08 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.09 |
| SVM RNA-class probability | 0.987755 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
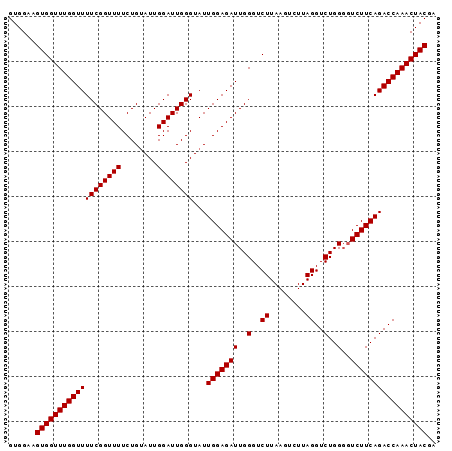
>3L_DroMel_CAF1 11380235 95 + 23771897 GUGGAAGUGGUUUGGUUUUCGGUUUUCUGUAUUGGAUUGGGUAUUGGAGAUUGGGUCUUAAGUCUUAGGUCUGGGGUCUUCAGACCAAACUACAA ......(((((((((((((((((((........))))))))...((((((((((..((........))..))..))))))))))))))))))).. ( -29.50) >DroSec_CAF1 100246 95 + 1 GUGGAAGUGGUUUGGUUUUCGGUUUUCUGCAUUGGAUUGGGUAUUGGAGAUUGGGUCUUAAGUCUUAGGUCUGGUUUCUUCAGACCAAACUACGA ......(((((((((((((((((((........))))))).....((((((..(..((........))..)..))))))..)))))))))))).. ( -30.80) >DroSim_CAF1 112696 95 + 1 GUGGAAGUGGUUUGGUUUUCGGUUUUCUGUAUUGGAUUGGGUAUUGGAGAUUGGGUCUUAAGUCUUAGGUCUGGGGUCUUCAGACCAAACUACGA ......(((((((((((((((((((........))))))))...((((((((((..((........))..))..))))))))))))))))))).. ( -29.50) >consensus GUGGAAGUGGUUUGGUUUUCGGUUUUCUGUAUUGGAUUGGGUAUUGGAGAUUGGGUCUUAAGUCUUAGGUCUGGGGUCUUCAGACCAAACUACGA ......(((((((((((((((((((........))))))))...(((((((..(..((........))..)..)..))))))))))))))))).. (-29.00 = -29.00 + 0.00)

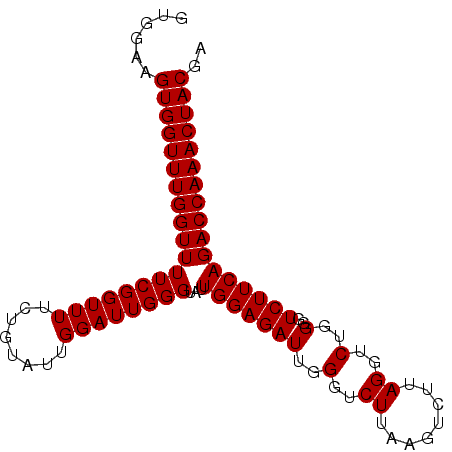
| Location | 11,380,235 – 11,380,330 |
|---|---|
| Length | 95 |
| Sequences | 3 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 97.19 |
| Mean single sequence MFE | -11.63 |
| Consensus MFE | -11.42 |
| Energy contribution | -11.20 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.584608 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11380235 95 - 23771897 UUGUAGUUUGGUCUGAAGACCCCAGACCUAAGACUUAAGACCCAAUCUCCAAUACCCAAUCCAAUACAGAAAACCGAAAACCAAACCACUUCCAC (((((((((((((((.......))))))..))))...(((.....)))................))))).......................... ( -11.30) >DroSec_CAF1 100246 95 - 1 UCGUAGUUUGGUCUGAAGAAACCAGACCUAAGACUUAAGACCCAAUCUCCAAUACCCAAUCCAAUGCAGAAAACCGAAAACCAAACCACUUCCAC ((..(((((((((((.......))))))..)))))...))....................................................... ( -11.80) >DroSim_CAF1 112696 95 - 1 UCGUAGUUUGGUCUGAAGACCCCAGACCUAAGACUUAAGACCCAAUCUCCAAUACCCAAUCCAAUACAGAAAACCGAAAACCAAACCACUUCCAC ((..(((((((((((.......))))))..)))))...))....................................................... ( -11.80) >consensus UCGUAGUUUGGUCUGAAGACCCCAGACCUAAGACUUAAGACCCAAUCUCCAAUACCCAAUCCAAUACAGAAAACCGAAAACCAAACCACUUCCAC ((..(((((((((((.......))))))..)))))...))....................................................... (-11.42 = -11.20 + -0.22)

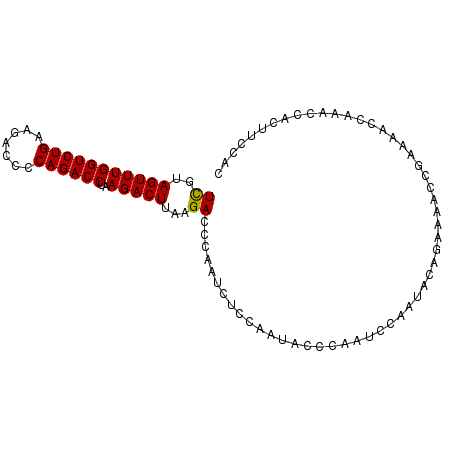
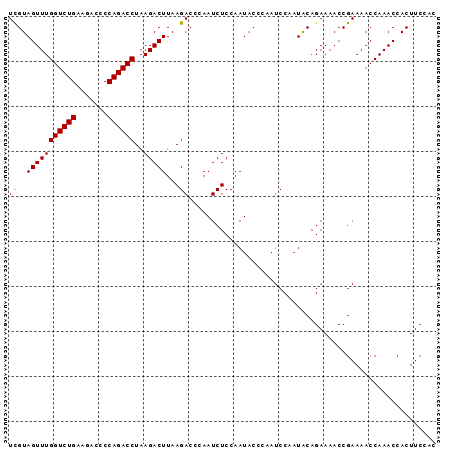
| Location | 11,380,300 – 11,380,399 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 82.27 |
| Mean single sequence MFE | -26.23 |
| Consensus MFE | -13.36 |
| Energy contribution | -13.12 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.17 |
| Mean z-score | -3.06 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.752577 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
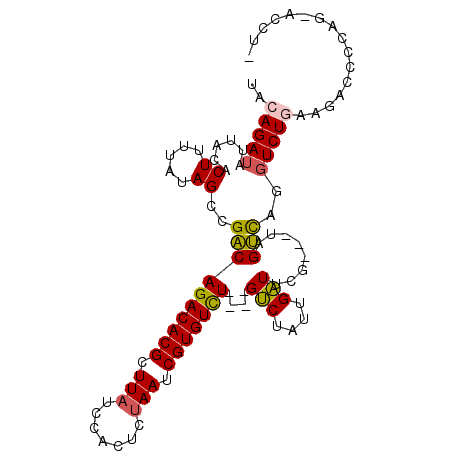
>3L_DroMel_CAF1 11380300 99 - 23771897 UACAGAUAUUACACUUUUAUAGCCGACAGACACGCUUAUCCACUCUAAUCGUGUCU----GUCUAUUGAUUCGUUGUAGUUUGGUCUGAAGACCCCAG-ACCUA .......((((((...........((((((((((.(((.......))).)))))))----)))...........))))))..((((((.......)))-))).. ( -27.85) >DroPse_CAF1 112919 99 - 1 UACAGAUAUUACACUUUUAUAGCCGACAGACACGCUUAUCCACUCUAAUCGUGUUU----GGCUGUCGCUCCGCCGUUGUCUUGUCUAUUGAUUCGUGGGCCC- ..................((((((((...(((((.(((.......))).)))))))----)))))).((.((((....(((.........)))..))))))..- ( -21.50) >DroSec_CAF1 100311 99 - 1 UACAGAUAUUACACUUUUAUAGCCGACAGACACGCUUAUCCACUCUAAUCGUGUCU----GUCUAUUGAUUCGUCGUAGUUUGGUCUGAAGAAACCAG-ACCUA .......(((((............((((((((((.(((.......))).)))))))----)))....((....)))))))..((((((.......)))-))).. ( -27.80) >DroEre_CAF1 106957 96 - 1 UACAGAUAUUACACUUUUAUAGCAGACAGACACGCUUAUCCACUCUAAUCGUGUCU----GUCUAUUGAUUCG---UAGUCAGGUCUGAAGACCUCGG-ACCUC ....(((..(((.((.....)).(((((((((((.(((.......))).)))))))----))))........)---)))))((((((((.....))))-)))). ( -29.90) >DroYak_CAF1 95523 96 - 1 UACAGAUAUUACACUUUUAUAGCCGACAGACACGCUUAUCCACUCUAAUCGUGUCU----GUCUAUUGAUUCG---UAGUCAGCUCUGAAGACCUCAA-AACCC .............((((...(((.((((((((((.(((.......))).)))))))----)))...((((...---..)))))))..)))).......-..... ( -23.30) >DroAna_CAF1 92353 91 - 1 UACAGAUAUUACACUUUUAUAGCCGGCAGACACGCUUAUCCACUCAAAUCGUGUCUGCCCGUCUAUUGAUUCG---UGGCCAGGUCUCAAGACC---------- ...........(((.((.(((((.((((((((((.((.........)).)))))))))).).)))).))...)---))....((((....))))---------- ( -27.00) >consensus UACAGAUAUUACACUUUUAUAGCCGACAGACACGCUUAUCCACUCUAAUCGUGUCU____GUCUAUUGAUUCG___UAGUCAGGUCUGAAGACCCCAG_ACCU_ ..(((((......((.....))..((((((((((.(((.......))).)))))))....(((....)))........)))..)))))................ (-13.36 = -13.12 + -0.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:10:09 2006