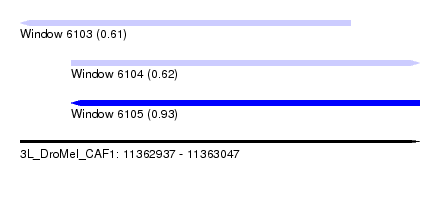
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 11,362,937 – 11,363,047 |
| Length | 110 |
| Max. P | 0.931675 |

| Location | 11,362,937 – 11,363,028 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 79.88 |
| Mean single sequence MFE | -21.52 |
| Consensus MFE | -19.03 |
| Energy contribution | -19.20 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.612811 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11362937 91 - 23771897 AAAUGCUAAUUUAUGGGCAUGUGUUGCCAUUUGGUUGCCGUUGGGUAAUUUUAGUGCCCACCAAAACACAGA-ACACA----------CAGAAAACACACAG---------------- .............(((((((..((((((...(((...)))...))))))....)))))))............-.....----------..............---------------- ( -20.50) >DroPse_CAF1 95150 106 - 1 AAAUGCUAAUUUAUGGGCAUGUGUUGCCAUUUGGUUGCCGUUGGGUAAUUUUAGUGGCCACCGAAACACACAC--ACA----------CACACAACAUACAGGGGAGGGGUGGGAGAG ..(((((........)))))(((((((((((.(((((((....)))))))..))))))......)))))...(--((.----------..(.(..(.....)..).)..)))...... ( -25.20) >DroEre_CAF1 87730 89 - 1 AAAUGCUAAUUUAUGGGCAUGUGUUGCCAUUUGGUUGCCGUUGGGUAAUUUCAGUGCCCACCAAAACACAGA-ACACA------------CAAAACACACAG---------------- .............(((((((..((((((...(((...)))...))))))....)))))))............-.....------------............---------------- ( -20.50) >DroWil_CAF1 30986 89 - 1 AAAUGCUAAUUUAUGGGCAUGUGUUGCCAUUUGGUUGCCGUUGGGUAAUUUUUGUGCCCACCAAAA-ACAGA-AAAAA-----------AGAAAUGAAACGG---------------- .............((((((((.((((((...(((...)))...))))))...))))))))......-.....-.....-----------.............---------------- ( -20.10) >DroYak_CAF1 77769 80 - 1 AAAUGCUAAUUUAUGGGCAUGUGUUGCCAUUUGGUUGCCGUUGGGUAAUUUUAGUGCCCACCAAAACACAGC-AC---------------------ACACAG---------------- ...((((..(((.(((((((..((((((...(((...)))...))))))....)))))))..)))....)))-).---------------------......---------------- ( -22.30) >DroAna_CAF1 74012 99 - 1 AAAUGCUAAUUUAUGGGCAUGUGUUGCCAUUUGGUUGCCGUUGGGUAAUUUUAGUGCCCACCAAAACACAAA-ACACAAAAUACAAAACACCAAACAUAC------------------ .............(((((((..((((((...(((...)))...))))))....)))))))............-...........................------------------ ( -20.50) >consensus AAAUGCUAAUUUAUGGGCAUGUGUUGCCAUUUGGUUGCCGUUGGGUAAUUUUAGUGCCCACCAAAACACAGA_ACACA___________ACAAAACACACAG________________ .............(((((((..((((((...(((...)))...))))))....))))))).......................................................... (-19.03 = -19.20 + 0.17)

| Location | 11,362,951 – 11,363,047 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 89.08 |
| Mean single sequence MFE | -19.12 |
| Consensus MFE | -17.25 |
| Energy contribution | -17.45 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.623048 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11362951 96 + 23771897 -UGUGUUCUGUGUUUUGGUGGGCACUAAAAUUACCCAACGGCAACCAAAUGGCAACACAUGCCCAUAAAUUAGCAUUUAGAAAAA-----------AAGGAGGAAAAC -....(((((((((...(((((((...............(....)....((....))..))))))).....)))))..))))...-----------............ ( -19.20) >DroSim_CAF1 92921 96 + 1 -UGUGUUCUGUGUUUUGGUGGGCACUAAAAUUACCCAACGGCAACCAAAUGGCAACACAUGCCCAUAAAUUAGCAUUUAGAAAAA-----------AAGGAGGAAACC -....(((((((((...(((((((...............(....)....((....))..))))))).....)))))..))))...-----------.....(....). ( -20.00) >DroEre_CAF1 87742 96 + 1 -UGUGUUCUGUGUUUUGGUGGGCACUGAAAUUACCCAACGGCAACCAAAUGGCAACACAUGCCCAUAAAUUAGCAUUUAGAAAAA-----------AAAGAGGAAAAC -....(((((((((...(((((((.((............(....)....((....))))))))))).....)))))..))))...-----------............ ( -19.90) >DroWil_CAF1 30999 95 + 1 -UUUUUUCUGU-UUUUGGUGGGCACAAAAAUUACCCAACGGCAACCAAAUGGCAACACAUGCCCAUAAAUUAGCAUUUAGAAAAA-----------AACAACAACAAU -((((((((((-(....(((((((...............(....)....((....))..))))))).....)))....)))))))-----------)........... ( -19.20) >DroYak_CAF1 77775 98 + 1 ----GUGCUGUGUUUUGGUGGGCACUAAAAUUACCCAACGGCAACCAAAUGGCAACACAUGCCCAUAAAUUAGCAUUUAGAAAAA------AAGAAAAAGAGGAAAAC ----((((((...((((.((((((...............(....)....((....))..)))))))))).)))))).........------................. ( -19.30) >DroAna_CAF1 74033 108 + 1 UUGUGUUUUGUGUUUUGGUGGGCACUAAAAUUACCCAACGGCAACCAAAUGGCAACACAUGCCCAUAAAUUAGCAUUUAGAAAAAAGAGAAAAAAAAAAGAGGAAAAC .....((..(((((...(((((((...............(....)....((....))..))))))).....)))))..))............................ ( -17.10) >consensus _UGUGUUCUGUGUUUUGGUGGGCACUAAAAUUACCCAACGGCAACCAAAUGGCAACACAUGCCCAUAAAUUAGCAUUUAGAAAAA___________AAAGAGGAAAAC .....(((((((((...(((((((...............(....)....((....))..))))))).....)))))..)))).......................... (-17.25 = -17.45 + 0.20)


| Location | 11,362,951 – 11,363,047 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 89.08 |
| Mean single sequence MFE | -22.95 |
| Consensus MFE | -20.52 |
| Energy contribution | -20.52 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.931675 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11362951 96 - 23771897 GUUUUCCUCCUU-----------UUUUUCUAAAUGCUAAUUUAUGGGCAUGUGUUGCCAUUUGGUUGCCGUUGGGUAAUUUUAGUGCCCACCAAAACACAGAACACA- (((((.......-----------......(((((....)))))(((((((..((((((...(((...)))...))))))....))))))).........)))))...- ( -22.70) >DroSim_CAF1 92921 96 - 1 GGUUUCCUCCUU-----------UUUUUCUAAAUGCUAAUUUAUGGGCAUGUGUUGCCAUUUGGUUGCCGUUGGGUAAUUUUAGUGCCCACCAAAACACAGAACACA- (((.........-----------......(((((....))))).((((((..((((((...(((...)))...))))))....)))))))))...............- ( -22.50) >DroEre_CAF1 87742 96 - 1 GUUUUCCUCUUU-----------UUUUUCUAAAUGCUAAUUUAUGGGCAUGUGUUGCCAUUUGGUUGCCGUUGGGUAAUUUCAGUGCCCACCAAAACACAGAACACA- (((((.......-----------......(((((....)))))(((((((..((((((...(((...)))...))))))....))))))).........)))))...- ( -22.70) >DroWil_CAF1 30999 95 - 1 AUUGUUGUUGUU-----------UUUUUCUAAAUGCUAAUUUAUGGGCAUGUGUUGCCAUUUGGUUGCCGUUGGGUAAUUUUUGUGCCCACCAAAA-ACAGAAAAAA- .......(((((-----------(((...(((((....)))))((((((((.((((((...(((...)))...))))))...))))))))..))))-))))......- ( -24.60) >DroYak_CAF1 77775 98 - 1 GUUUUCCUCUUUUUCUU------UUUUUCUAAAUGCUAAUUUAUGGGCAUGUGUUGCCAUUUGGUUGCCGUUGGGUAAUUUUAGUGCCCACCAAAACACAGCAC---- (((((............------......(((((....)))))(((((((..((((((...(((...)))...))))))....)))))))..))))).......---- ( -22.60) >DroAna_CAF1 74033 108 - 1 GUUUUCCUCUUUUUUUUUUCUCUUUUUUCUAAAUGCUAAUUUAUGGGCAUGUGUUGCCAUUUGGUUGCCGUUGGGUAAUUUUAGUGCCCACCAAAACACAAAACACAA (((((........................(((((....)))))(((((((..((((((...(((...)))...))))))....)))))))..)))))........... ( -22.60) >consensus GUUUUCCUCUUU___________UUUUUCUAAAUGCUAAUUUAUGGGCAUGUGUUGCCAUUUGGUUGCCGUUGGGUAAUUUUAGUGCCCACCAAAACACAGAACACA_ .............................(((((....)))))(((((((..((((((...(((...)))...))))))....))))))).................. (-20.52 = -20.52 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:10:03 2006