
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 11,345,193 – 11,345,363 |
| Length | 170 |
| Max. P | 0.999934 |

| Location | 11,345,193 – 11,345,307 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 90.15 |
| Mean single sequence MFE | -41.15 |
| Consensus MFE | -33.48 |
| Energy contribution | -33.48 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.13 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.923297 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11345193 114 + 23771897 AACUCCGGGAAACCGUAGCUCCUGUCCUCCACUAUCCUGACUAUCCUGUCCGUCCUGCCAGUUAGCCAGUUGGCCAGUUGGCCAUUUGGCUCUUUGGCCGUUUGGCUGUCAGGC ..((.(((....))).)).................((((((......(.(((..(.(((((..((((((.(((((....))))).))))))..))))).)..)))).)))))). ( -46.90) >DroSec_CAF1 65735 107 + 1 AACUCCGGGAGCUUGCAGCUCCUGUCCUCCACUAUCCUGACUAUCCUAUCCGUCCU-------UGCCAGUUGGCCAGUUGGCCAUUUCGCUGUUUGGCCGUUUGGCUGUCAGGC .....(((((((.....)))))))...........((((((...............-------.(((((.(((((((..(((......)))..))))))).))))).)))))). ( -40.53) >DroSim_CAF1 74155 107 + 1 AACUCCGGGAAGCCGCAGCUCCUGUCCUCCACUAUCCUGACUAUCCUAUCCGUCCU-------UGCCAGUUGGCCAGUUGGCCAUUUCGCUGUUUGGCCGUUUGGCUGUCAGGC .....(((((.((....)))))))...........((((((...............-------.(((((.(((((((..(((......)))..))))))).))))).)))))). ( -36.03) >consensus AACUCCGGGAAACCGCAGCUCCUGUCCUCCACUAUCCUGACUAUCCUAUCCGUCCU_______UGCCAGUUGGCCAGUUGGCCAUUUCGCUGUUUGGCCGUUUGGCUGUCAGGC .....(((((.........)))))...........((((((.......................(((((.(((((((..(((......)))..))))))).))))).)))))). (-33.48 = -33.48 + -0.00)

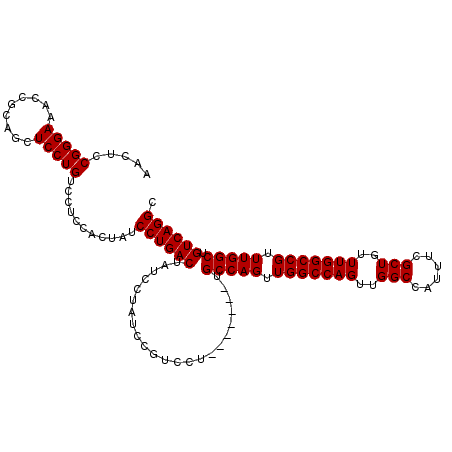

| Location | 11,345,233 – 11,345,342 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 87.58 |
| Mean single sequence MFE | -43.38 |
| Consensus MFE | -39.44 |
| Energy contribution | -39.88 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.13 |
| Mean z-score | -3.72 |
| Structure conservation index | 0.91 |
| SVM decision value | 4.66 |
| SVM RNA-class probability | 0.999934 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11345233 109 + 23771897 -CUAUCCUGUCCGUCCUGCCAGUUAGCCAGUUGGCCAGUUGGCCAUUUGGCUCUUUGGCCGUUUGGCUGUCAGGCAGCUGAAUAAUUUAGCUGCGAUAAUAAGCGGCUGC -..............(((((...(((((((.(((((((..((((....))))..))))))).)))))))...)))))..........(((((((........))))))). ( -48.30) >DroSec_CAF1 65775 102 + 1 -CUAUCCUAUCCGUCCU-------UGCCAGUUGGCCAGUUGGCCAUUUCGCUGUUUGGCCGUUUGGCUGUCAGGCAGCUGAAUAAUUUAGCUGCGAUAAUAAGCGGCUGU -.........((((...-------.(((((.(((((((..(((......)))..))))))).)))))((((..((((((((.....))))))))))))....)))).... ( -38.60) >DroSim_CAF1 74195 102 + 1 -CUAUCCUAUCCGUCCU-------UGCCAGUUGGCCAGUUGGCCAUUUCGCUGUUUGGCCGUUUGGCUGUCAGGCAGCUGAAUAAUUUAGCUGCGAUAAUAAGCGGCUGU -.........((((...-------.(((((.(((((((..(((......)))..))))))).)))))((((..((((((((.....))))))))))))....)))).... ( -38.60) >DroEre_CAF1 70195 108 + 1 --UGUCCUGUCCGUCCUGCUAGUUGGCCAUUUGGCUGUUUGGCCGUUUGGCUGUUUGGCCGUUUGGCUGUCAGGCAGCUGAAUAAUUUAGCUGCGAUAAUAAGCGGCUGC --......................(((((..(((((....)))))..)))))....(((((((((..((((..((((((((.....)))))))))))).))))))))).. ( -42.60) >DroYak_CAF1 59333 110 + 1 CCUGUCCUAUCCGUAAUGCUAGUUGGCCAUUUGGCUGUUUGGCCAUUUGGCCAUUUGGCCGUUUGGCUGUCAGGCAGCUGAAUAAUUUAGCCGCGAUAAUAAGCGGCUGU .................(((((.((((((..(((((....)))))..)))))).))))).(((..((((.....))))..)))....(((((((........))))))). ( -48.80) >consensus _CUAUCCUAUCCGUCCUGC_AGUUGGCCAGUUGGCCAGUUGGCCAUUUGGCUGUUUGGCCGUUUGGCUGUCAGGCAGCUGAAUAAUUUAGCUGCGAUAAUAAGCGGCUGU ........................(((((..(((((....)))))..)))))....(((((((((..((((..((((((((.....)))))))))))).))))))))).. (-39.44 = -39.88 + 0.44)


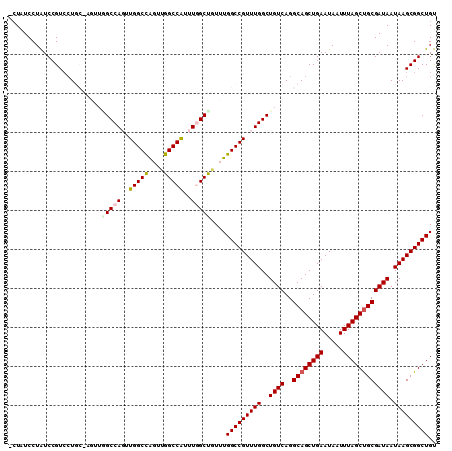
| Location | 11,345,233 – 11,345,342 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 87.58 |
| Mean single sequence MFE | -33.24 |
| Consensus MFE | -25.60 |
| Energy contribution | -26.44 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.965653 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11345233 109 - 23771897 GCAGCCGCUUAUUAUCGCAGCUAAAUUAUUCAGCUGCCUGACAGCCAAACGGCCAAAGAGCCAAAUGGCCAACUGGCCAACUGGCUAACUGGCAGGACGGACAGGAUAG- ((....))....((((((((((.........))))))(((....((...(.((((...(((((..(((((....)))))..)))))...)))).)...)).))))))).- ( -39.70) >DroSec_CAF1 65775 102 - 1 ACAGCCGCUUAUUAUCGCAGCUAAAUUAUUCAGCUGCCUGACAGCCAAACGGCCAAACAGCGAAAUGGCCAACUGGCCAACUGGCA-------AGGACGGAUAGGAUAG- ....((((((......((((((.........))))))......((((...(((((....((......))....)))))...)))).-------))).))).........- ( -30.00) >DroSim_CAF1 74195 102 - 1 ACAGCCGCUUAUUAUCGCAGCUAAAUUAUUCAGCUGCCUGACAGCCAAACGGCCAAACAGCGAAAUGGCCAACUGGCCAACUGGCA-------AGGACGGAUAGGAUAG- ....((((((......((((((.........))))))......((((...(((((....((......))....)))))...)))).-------))).))).........- ( -30.00) >DroEre_CAF1 70195 108 - 1 GCAGCCGCUUAUUAUCGCAGCUAAAUUAUUCAGCUGCCUGACAGCCAAACGGCCAAACAGCCAAACGGCCAAACAGCCAAAUGGCCAACUAGCAGGACGGACAGGACA-- ...(((((((......((((((.........))))))......((((...(((......)))....(((......)))...))))........))).))).)......-- ( -27.90) >DroYak_CAF1 59333 110 - 1 ACAGCCGCUUAUUAUCGCGGCUAAAUUAUUCAGCUGCCUGACAGCCAAACGGCCAAAUGGCCAAAUGGCCAAACAGCCAAAUGGCCAACUAGCAUUACGGAUAGGACAGG ..((((((........))))))...........(((((((.(.(((....)))....((((((..((((......))))..))))))............).)))).))). ( -38.60) >consensus ACAGCCGCUUAUUAUCGCAGCUAAAUUAUUCAGCUGCCUGACAGCCAAACGGCCAAACAGCCAAAUGGCCAACUGGCCAACUGGCAAACU_GCAGGACGGAUAGGAUAG_ ....((..........((((((.........))))))(((...(((....)))......((((..(((((....)))))..))))............)))...))..... (-25.60 = -26.44 + 0.84)


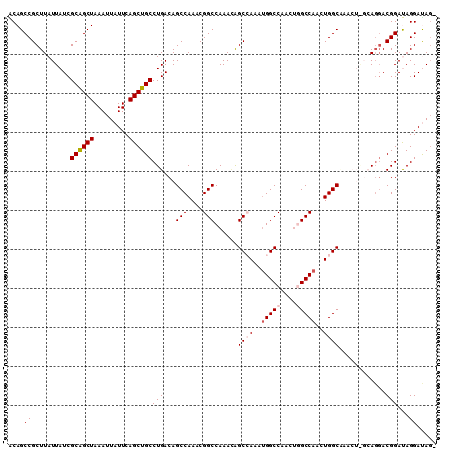
| Location | 11,345,271 – 11,345,363 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 85.81 |
| Mean single sequence MFE | -35.70 |
| Consensus MFE | -26.80 |
| Energy contribution | -27.53 |
| Covariance contribution | 0.74 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.59 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.895682 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11345271 92 + 23771897 UGGCCAUUUGGCUCUUUGGCCGUUUGGCUGUCAGGCAGCUGAAUAAUUUAGCUGCGAUAAUAAGCGGCUGCAGUGCCAGUGUCCACUGGUUG (((.((((.(((.((.((((((((((..((((..((((((((.....)))))))))))).)))))))))).)).))))))).)))....... ( -40.70) >DroSec_CAF1 65806 87 + 1 UGGCCAUUUCGCUGUUUGGCCGUUUGGCUGUCAGGCAGCUGAAUAAUUUAGCUGCGAUAAUAAGCGGCUGUAGUGC-AGUGUCCACUG---- (((.((((.(((((..((((((((((..((((..((((((((.....)))))))))))).))))))))))))))).-)))).)))...---- ( -37.10) >DroSim_CAF1 74226 87 + 1 UGGCCAUUUCGCUGUUUGGCCGUUUGGCUGUCAGGCAGCUGAAUAAUUUAGCUGCGAUAAUAAGCGGCUGUAGUGC-AGUGUCCACUG---- (((.((((.(((((..((((((((((..((((..((((((((.....)))))))))))).))))))))))))))).-)))).)))...---- ( -37.10) >DroEre_CAF1 70232 87 + 1 UGGCCGUUUGGCUGUUUGGCCGUUUGGCUGUCAGGCAGCUGAAUAAUUUAGCUGCGAUAAUAAGCGGCUGCAGUGC-AGUGUCCACUG---- .(((((((((.(((..(((((....))))).)))((((((((.....)))))))).....))))))))).(((((.-......)))))---- ( -35.60) >DroYak_CAF1 59372 91 + 1 UGGCCAUUUGGCCAUUUGGCCGUUUGGCUGUCAGGCAGCUGAAUAAUUUAGCCGCGAUAAUAAGCGGCUGUAGUGC-AGUGUCCACUCGUCC .(((((..(((((....)))))..)))))....((..((((....((((((((((........))))))).))).)-)))..))........ ( -38.20) >DroAna_CAF1 57808 71 + 1 ------------CAGUUG---GUCCGGCUGUCAGGCUGCUGAAUAAUUUAGCUGCGAUAAUAAGCGGCUGUAGC-C-AGUGUCCACUG---- ------------((((.(---(.(((((((((((....))))......(((((((........)))))))))))-)-.).).))))))---- ( -25.50) >consensus UGGCCAUUUGGCUGUUUGGCCGUUUGGCUGUCAGGCAGCUGAAUAAUUUAGCUGCGAUAAUAAGCGGCUGUAGUGC_AGUGUCCACUG____ .(((((..........)))))(((((((((.....)))))))))....(((((((........)))))))(((((........))))).... (-26.80 = -27.53 + 0.74)


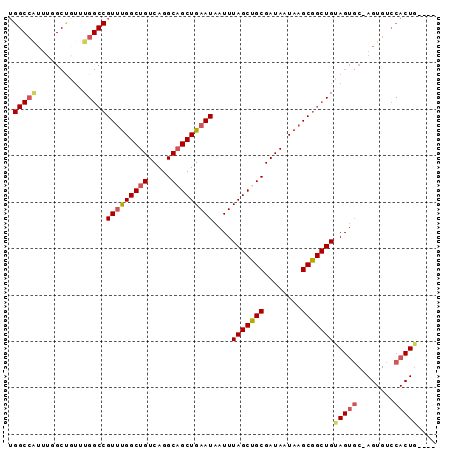
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:09:59 2006