
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 11,333,772 – 11,333,862 |
| Length | 90 |
| Max. P | 0.996087 |

| Location | 11,333,772 – 11,333,862 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 76.54 |
| Mean single sequence MFE | -34.27 |
| Consensus MFE | -20.36 |
| Energy contribution | -22.05 |
| Covariance contribution | 1.69 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.59 |
| SVM decision value | 2.65 |
| SVM RNA-class probability | 0.996087 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
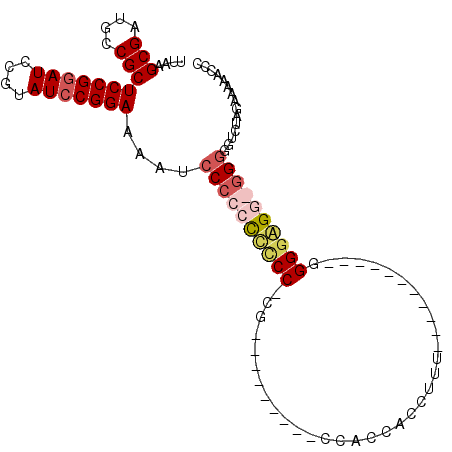
>3L_DroMel_CAF1 11333772 90 + 23771897 UUAAGCGAUGCCGCUCCGGAUCCGUAUCCGGAAAAUCCCUCCCCCC-CACUCCAUCCACCAUCACCUUU-----------GGGAAGG-UG---GCUAGAAAAACCC ....(((....)))(((((((....)))))))..............-...((...(((((.((.((...-----------)))).))-))---)...))....... ( -22.90) >DroSim_CAF1 61585 90 + 1 UUAAGCGAUGCCGCUCCGGAUCCGUAUCCGGAAAACCCCCCCUCCC-CGCUG---CCAACUCCACCUUU-----------GGGGAGG-GGGAGUCUAGAAAAACCC ....(((....)))(((((((....)))))))..((.(((((((((-((..(---..........)..)-----------)))))))-))).))............ ( -36.90) >DroEre_CAF1 58310 84 + 1 UUAAGCGAUGCCGCUCCGUAUCCGUAUCCGGAAAAUCCCCCUCCCC-UG---------CCACCACCUUU-----------GGGGGGUGGGGGG-CUAGAAAAACCC ...((((....)))).....((((....))))...((((((.((((-..---------(((.......)-----------)))))).))))))-............ ( -29.90) >DroYak_CAF1 46991 97 + 1 UUAAGCGAUGCCGCUCCGGAUCCGUAUCCGGAAAAUCCCCCCCUCCACG---------CCACCACCUUUCAGGGGGGAGUGGGGGGUGGGGGGUCUAGAAAAACCC ....(((....)))(((((((....)))))))....((((((((((((.---------((.((.((.....)))))).)))))))).))))(((........))). ( -47.40) >consensus UUAAGCGAUGCCGCUCCGGAUCCGUAUCCGGAAAAUCCCCCCCCCC_CG_________CCACCACCUUU___________GGGGAGG_GGGGGUCUAGAAAAACCC ....(((....)))(((((((....)))))))....((((((((((...................................))))))))))............... (-20.36 = -22.05 + 1.69)


| Location | 11,333,772 – 11,333,862 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 76.54 |
| Mean single sequence MFE | -39.27 |
| Consensus MFE | -21.04 |
| Energy contribution | -22.72 |
| Covariance contribution | 1.69 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.886453 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11333772 90 - 23771897 GGGUUUUUCUAGC---CA-CCUUCCC-----------AAAGGUGAUGGUGGAUGGAGUG-GGGGGGAGGGAUUUUCCGGAUACGGAUCCGGAGCGGCAUCGCUUAA ((((((.((((.(---((-((.((((-----------...)).)).))))).))))(((-.((..((....))..))...))))))))).(((((....))))).. ( -31.00) >DroSim_CAF1 61585 90 - 1 GGGUUUUUCUAGACUCCC-CCUCCCC-----------AAAGGUGGAGUUGG---CAGCG-GGGAGGGGGGGUUUUCCGGAUACGGAUCCGGAGCGGCAUCGCUUAA ..........((((((((-(((((((-----------...(.((.......---)).))-))))))))))))))(((((((....)))))))(((....))).... ( -43.70) >DroEre_CAF1 58310 84 - 1 GGGUUUUUCUAG-CCCCCCACCCCCC-----------AAAGGUGGUGG---------CA-GGGGAGGGGGAUUUUCCGGAUACGGAUACGGAGCGGCAUCGCUUAA .....(..((..-(((((((((((..-----------...)).)))))---------..-))))..))..)...((((....))))....(((((....))))).. ( -33.00) >DroYak_CAF1 46991 97 - 1 GGGUUUUUCUAGACCCCCCACCCCCCACUCCCCCCUGAAAGGUGGUGG---------CGUGGAGGGGGGGAUUUUCCGGAUACGGAUCCGGAGCGGCAUCGCUUAA ((((((....))))))(((.((((((((.((((((.....)).)).))---------.)))).)))))))....(((((((....)))))))(((....))).... ( -49.40) >consensus GGGUUUUUCUAGACCCCC_ACCCCCC___________AAAGGUGGUGG_________CG_GGGGGGGGGGAUUUUCCGGAUACGGAUCCGGAGCGGCAUCGCUUAA ...............((((((((((...................................))))))))))....(((((((....)))))))(((....))).... (-21.04 = -22.72 + 1.69)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:09:54 2006