
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
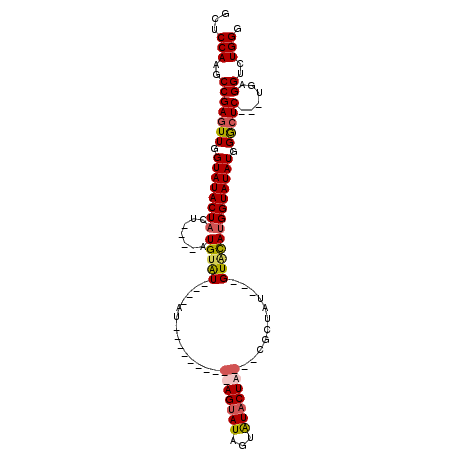
| Location | 11,324,399 – 11,324,491 |
| Length | 92 |
| Max. P | 0.998949 |

| Location | 11,324,399 – 11,324,491 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 76.54 |
| Mean single sequence MFE | -34.10 |
| Consensus MFE | -16.75 |
| Energy contribution | -17.75 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.17 |
| Mean z-score | -4.20 |
| Structure conservation index | 0.49 |
| SVM decision value | 3.30 |
| SVM RNA-class probability | 0.998949 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
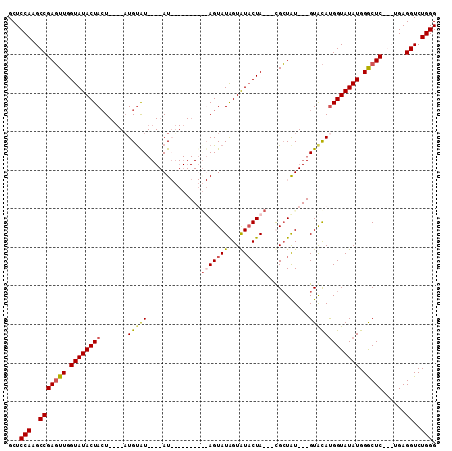
>3L_DroMel_CAF1 11324399 92 + 23771897 GCUCCAAGCCGAGUUGGUAUACUAAU----AUGCAU----AUAU-----UGUAGUAUAGUAUACUACUAUGCUAU---GUACAUGGUAUAUGGGCUC---CGAGGUCUGGG ...(((..(((((((.((((((((..----.(((((----((((-----.(((((((...))))))).))).)))---)))..)))))))).)))))---...))..))). ( -33.30) >DroSec_CAF1 45060 87 + 1 GCUCCAAGCCGAGUUGGUAUACUACU----AUGUAU----AU----------AGUAUAGUAUACUA---CGCUAU---GUACAUGGUAUAUGGACUCGGCUGAGGUCUGGG .(((..(((((((((.(((((((...----((((((----((----------((..(((....)))---..))))---))))))))))))).))))))))))))....... ( -42.10) >DroSim_CAF1 52163 87 + 1 GCUCCAAGCCGAGUUGGUAUACUACU----AUGUAU----AU----------AGUAUAGUAUACUA---CGCUAU---GUACACGGUAUAUGGGCUCAGCUGAGGUCUGGG .(((..(((.(((((.(((((((...----.(((((----((----------((..(((....)))---..))))---))))).))))))).))))).))))))....... ( -33.90) >DroYak_CAF1 37358 105 + 1 GCUCCAAGCCGACUUGGUAUACUACUUACUUUGUGUAUAUAUAGGUAUAUAUAGUAUGGUGUUCUA---UGGUGUAUGGUGUAUGGUAUAUGGGCUC---UGAGGUCUGGG .(((..((((.(..(.((((((((..(((.((((((((((....))))))))))(((((....)))---)))))..)))))))).)....).)))).---.)))....... ( -27.10) >consensus GCUCCAAGCCGAGUUGGUAUACUACU____AUGUAU____AU__________AGUAUAGUAUACUA___CGCUAU___GUACAUGGUAUAUGGGCUC___UGAGGUCUGGG ...(((..(((((((.((((((((.......(((((..............(((((((...)))))))...........))))))))))))).)))))......))..))). (-16.75 = -17.75 + 1.00)

| Location | 11,324,399 – 11,324,491 |
|---|---|
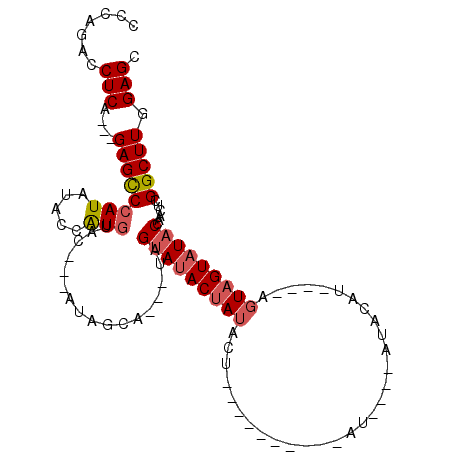
| Length | 92 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 76.54 |
| Mean single sequence MFE | -22.68 |
| Consensus MFE | -9.34 |
| Energy contribution | -10.21 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.15 |
| Structure conservation index | 0.41 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.913961 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11324399 92 - 23771897 CCCAGACCUCG---GAGCCCAUAUACCAUGUAC---AUAGCAUAGUAGUAUACUAUACUACA-----AUAU----AUGCAU----AUUAGUAUACCAACUCGGCUUGGAGC .......(((.---(((((....(((.((((..---...)))).)))((((((((.....((-----....----.))...----..))))))))......))))).))). ( -23.20) >DroSec_CAF1 45060 87 - 1 CCCAGACCUCAGCCGAGUCCAUAUACCAUGUAC---AUAGCG---UAGUAUACUAUACU----------AU----AUACAU----AGUAGUAUACCAACUCGGCUUGGAGC ......((..((((((((...(((((.(((((.---((((..---(((....)))..))----------))----.)))))----....)))))...)))))))).))... ( -28.50) >DroSim_CAF1 52163 87 - 1 CCCAGACCUCAGCUGAGCCCAUAUACCGUGUAC---AUAGCG---UAGUAUACUAUACU----------AU----AUACAU----AGUAGUAUACCAACUCGGCUUGGAGC ......((..(((((((....(((((.(((((.---((((..---(((....)))..))----------))----.)))))----....)))))....))))))).))... ( -24.80) >DroYak_CAF1 37358 105 - 1 CCCAGACCUCA---GAGCCCAUAUACCAUACACCAUACACCA---UAGAACACCAUACUAUAUAUACCUAUAUAUACACAAAGUAAGUAGUAUACCAAGUCGGCUUGGAGC .......(((.---(((((.((.....((((..(.(((....---..(.....)....(((((((....)))))))......))).)..)))).....)).))))).))). ( -14.20) >consensus CCCAGACCUCA___GAGCCCAUAUACCAUGUAC___AUAGCA___UAGUAUACUAUACU__________AU____AUACAU____AGUAGUAUACCAACUCGGCUUGGAGC .......(((....((((((((.....))).................(((((((((..............................)))))))))......))))).))). ( -9.34 = -10.21 + 0.87)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:09:46 2006