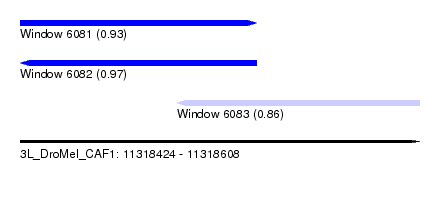
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 11,318,424 – 11,318,608 |
| Length | 184 |
| Max. P | 0.967473 |

| Location | 11,318,424 – 11,318,533 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 78.39 |
| Mean single sequence MFE | -25.42 |
| Consensus MFE | -9.88 |
| Energy contribution | -10.36 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.39 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.928989 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11318424 109 + 23771897 UUAUAGAACUUUCUAGUUUCACUAGCAAACUAAAAGAUACAUGUUUUGUAUCUUGAGG-------UAAGCA-ACCGUGAGUACUUCCAGGUACAUCUACCUAGAAACUGCUCAUCUC .....(((((....))))).....((..(((..((((((((.....))))))))..))-------)..)).-...(((((((.(((.(((((....))))).)))..)))))))... ( -28.50) >DroSec_CAF1 39226 109 + 1 UUAUAGAACUUUCUAAUUUAACUAGCAAACUAAAAGAUACAUUUUUUGUAUCUUAAGG-------UAAGCA-GCUGUGAGUACUUUCAGGUACAUCUACCUAGAUACUGCUCAUCUC ...((((....)))).........((..(((..((((((((.....))))))))..))-------)..)).-...(((((((...(((((((....))))).))...)))))))... ( -26.30) >DroSim_CAF1 46269 109 + 1 UUAUAGAACUUUCUCGUUUGACUAGCAAACUAGAAGAUACAUUUUUUAUAUCUUAAGG-------UAAGCA-GCUGUGAGUACUUCCAGGUACAUCUACCUAGAUACUGCUCAUCUC .........(((((.(((((.....))))).)))))...................(((-------(.((((-(......(((((....)))))((((....)))).))))).)))). ( -22.00) >DroEre_CAF1 43094 105 + 1 UUGUAGAACUUUCUAGAUACGCAGUUAAACU-----------ACGAUGUAUCUUAAGGCUCUAAGCAAGCA-ACUGAGCGUACUUUCAGGUACAUCUACCUAGAUACUGCUCAUCUC (((((((.((((..(((((((..((......-----------))..))))))).)))).)))..))))...-..((((((((((....)))))((((....))))...))))).... ( -26.60) >DroYak_CAF1 31030 117 + 1 UUAUGGAACUUUCCAGUUCUGCAGUCAAACUAAAAGAGGAAUAUGAUGUAUCUCAAGGCACUAAGUAAGCAAACUGAGCGUACUUCCAGGUACAUCUACCUAGAUACCGCUCAUCUC ...((((....))))(((.(((.............((((.((.....)).))))...((.....))..))))))((((((....((.(((((....))))).))...)))))).... ( -23.70) >consensus UUAUAGAACUUUCUAGUUUCACUAGCAAACUAAAAGAUACAUAUUUUGUAUCUUAAGG_______UAAGCA_ACUGUGAGUACUUCCAGGUACAUCUACCUAGAUACUGCUCAUCUC ...((((....))))................................((((((..(((.........((....))....(((((....))))).....))))))))).......... ( -9.88 = -10.36 + 0.48)

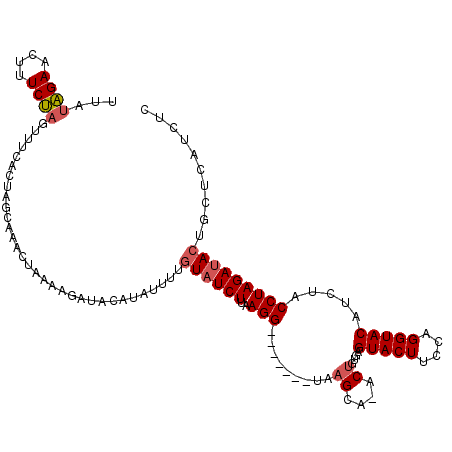
| Location | 11,318,424 – 11,318,533 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 78.39 |
| Mean single sequence MFE | -29.48 |
| Consensus MFE | -16.42 |
| Energy contribution | -17.34 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.967473 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11318424 109 - 23771897 GAGAUGAGCAGUUUCUAGGUAGAUGUACCUGGAAGUACUCACGGU-UGCUUA-------CCUCAAGAUACAAAACAUGUAUCUUUUAGUUUGCUAGUGAAACUAGAAAGUUCUAUAA (((.(((((((.(((((((((....)))))))))((....))..)-))))))-------.)))((((((((.....)))))))).(((..(.((((.....))))...)..)))... ( -31.50) >DroSec_CAF1 39226 109 - 1 GAGAUGAGCAGUAUCUAGGUAGAUGUACCUGAAAGUACUCACAGC-UGCUUA-------CCUUAAGAUACAAAAAAUGUAUCUUUUAGUUUGCUAGUUAAAUUAGAAAGUUCUAUAA (((.((((((((...((((((....))))))...((....)).))-))))))-------.)))..((((((.....))))))((((((((((.....)))))))))).......... ( -25.70) >DroSim_CAF1 46269 109 - 1 GAGAUGAGCAGUAUCUAGGUAGAUGUACCUGGAAGUACUCACAGC-UGCUUA-------CCUUAAGAUAUAAAAAAUGUAUCUUCUAGUUUGCUAGUCAAACGAGAAAGUUCUAUAA (((.((((((((.((((((((....)))))))).((....)).))-))))))-------.)))..((((((.....))))))((((.(((((.....))))).)))).......... ( -30.30) >DroEre_CAF1 43094 105 - 1 GAGAUGAGCAGUAUCUAGGUAGAUGUACCUGAAAGUACGCUCAGU-UGCUUGCUUAGAGCCUUAAGAUACAUCGU-----------AGUUUAACUGCGUAUCUAGAAAGUUCUACAA (((.(((((.((((.((((((....))))))...)))))))))..-..)))...((((((.((.((((((...((-----------((.....))))))))))..)).))))))... ( -29.30) >DroYak_CAF1 31030 117 - 1 GAGAUGAGCGGUAUCUAGGUAGAUGUACCUGGAAGUACGCUCAGUUUGCUUACUUAGUGCCUUGAGAUACAUCAUAUUCCUCUUUUAGUUUGACUGCAGAACUGGAAAGUUCCAUAA ...(((.(((((.((((((((....)))))))).(((..(((((..((((.....))))..))))).)))......................))))).(((((....)))))))).. ( -30.60) >consensus GAGAUGAGCAGUAUCUAGGUAGAUGUACCUGGAAGUACUCACAGU_UGCUUA_______CCUUAAGAUACAAAAAAUGUAUCUUUUAGUUUGCUAGUGAAACUAGAAAGUUCUAUAA .....((((.(((((((((((....)))))..(((((.........))))).............))))))............(((((((((.......))))))))).))))..... (-16.42 = -17.34 + 0.92)

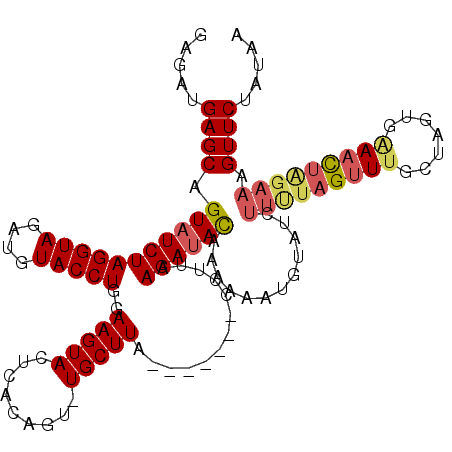
| Location | 11,318,496 – 11,318,608 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 94.09 |
| Mean single sequence MFE | -33.14 |
| Consensus MFE | -28.52 |
| Energy contribution | -28.92 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.856582 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11318496 112 - 23771897 ACUCACAGCCUCUGCCUGGCAUUGUUGUCAUAAAUUACACUUCAAUUGUUGCCCAGGUCGAAGCACUUGCCAUCGGAGAUGAGCAGUUUCUAGGUAGAUGUACCUGGAAGUA .......((.((.((((((((..((((...............))))...)).)))))).)).))..((((((((...)))).))))((((((((((....)))))))))).. ( -33.66) >DroSec_CAF1 39298 112 - 1 ACUCACAGCCUCUGCCUGGCAUUGUUGUCAUAAAUUACACUUCAAUUGUUGCCCAGGUGGAAGCACUUGCCAUCGGAGAUGAGCAGUAUCUAGGUAGAUGUACCUGAAAGUA .......((.((..(((((((..((((...............))))...)).)))))..)).))((((((((((...)))).))......((((((....)))))).)))). ( -31.16) >DroSim_CAF1 46341 112 - 1 ACUCACAGCCUCUGCCUGGCAUUGUUGUCAUAAAUUACACUUCAAUUGUUGCCCAGGUCGAAGCACUUGCCAUCGGAGAUGAGCAGUAUCUAGGUAGAUGUACCUGGAAGUA .......((.((.((((((((..((((...............))))...)).)))))).)).))(((.((((((...)))).))))).((((((((....)))))))).... ( -32.66) >DroEre_CAF1 43162 108 - 1 ACU----GCCUCUGCCUGGCAUUGUUGUCAUAAAUUACACUUCAAUUGUGGCCCAGGUCGGGGCACUUGCCAUCAGAGAUGAGCAGUAUCUAGGUAGAUGUACCUGAAAGUA ..(----(((((.((((((.......(((((((............))))))))))))).)))))).((((((((...)))).))))....((((((....))))))...... ( -33.71) >DroYak_CAF1 31110 108 - 1 ACU----GCCUCUGCCUGGCAUUGUUGUCAUAAAUUACACUUCAAUUGUUGCCCAGGUCGGGCCACUUGCCAUCGGAGAUGAGCGGUAUCUAGGUAGAUGUACCUGGAAGUA (((----(((((((..((((((((.(((........)))...)))..((.((((.....)))).)).))))).)))))....))))).((((((((....)))))))).... ( -34.50) >consensus ACUCACAGCCUCUGCCUGGCAUUGUUGUCAUAAAUUACACUUCAAUUGUUGCCCAGGUCGAAGCACUUGCCAUCGGAGAUGAGCAGUAUCUAGGUAGAUGUACCUGGAAGUA .......((.((..(((((((..((((...............))))...)).)))))..)).))(((.((((((...)))).))))).((((((((....)))))))).... (-28.52 = -28.92 + 0.40)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:09:42 2006