| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 11,301,572 – 11,301,666 |
| Length | 94 |
| Max. P | 0.983737 |

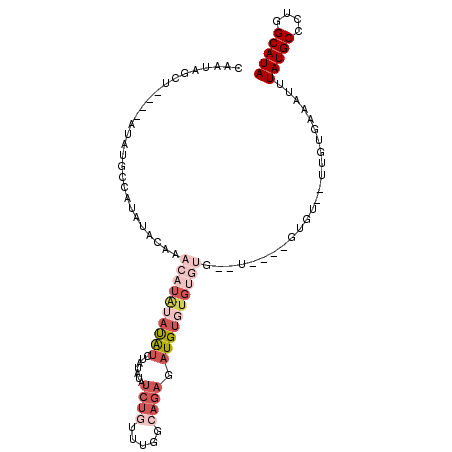
| Location | 11,301,572 – 11,301,666 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 66.00 |
| Mean single sequence MFE | -17.89 |
| Consensus MFE | -7.24 |
| Energy contribution | -7.44 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.47 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.40 |
| SVM decision value | 1.95 |
| SVM RNA-class probability | 0.983737 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11301572 94 + 23771897 UAUGCCAGGGCAUAAAUUUUACAA----------A--CACACACACAUCUCUGCCAUACAGAUAUAUAGAUAUAUAUGUUUGUAUAUAGCAUAUAUGUAGCUAUUG (((((....)))))..........----------.--.......((((.(.(((.(((((((((((((....)))))))))))))...))).).))))........ ( -21.40) >DroSec_CAF1 25710 92 + 1 UAUGCCAGGGCAUAAAUUUCACAG----------ACACACACACACAUCUCUGCCAAACAGAUAUAGAUAUGUAUAUGUUUGUAUAUGGCAUAU----AGCUAUUG (((((((.((((............----------.................))))..(((((((((........)))))))))...))))))).----........ ( -19.55) >DroSim_CAF1 29944 102 + 1 UAUGCCAGGGCAUAAAUUUCACAAAAACACAGACACACACACACACAUCUCUGCCAAACAGAAAUAUAGAUAUAUAUGUUUGUAUAUAGCAUAU----AGCUAUUG (((((....)))))..............((((((...............((((.....)))).(((((....)))))))))))..(((((....----.))))).. ( -18.40) >DroEre_CAF1 29303 82 + 1 UAUGCCAGGGCAUAAAUUUCACAA--ACAC--------------ACAUAUCUGC-AAAAAGAU--AUAGAUAUAUAAGUACCAGUAU-GUAUAU----AGCUAGUG (((((....)))))..........--...(--------------((.(((((..-....))))--)(((.((((((.(((....)))-.)))))----).)))))) ( -16.80) >DroPer_CAF1 32446 85 + 1 UAUGCCAGGGCAUAAAUUUCACGAACACACAC-------UCCACACA-------CUCCACACU--CCACACA-CCAUCCAUGGCCAUGGGGUUU----ACCUAUUG (((((....)))))..................-------........-------......(((--(((....-(((....)))...))))))..----........ ( -13.30) >consensus UAUGCCAGGGCAUAAAUUUCACAA__ACAC____A__CACACACACAUCUCUGCCAAACAGAUAUAUAGAUAUAUAUGUUUGUAUAUAGCAUAU____AGCUAUUG (((((....)))))........................................................((((((....))))))((((.........))))... ( -7.24 = -7.44 + 0.20)

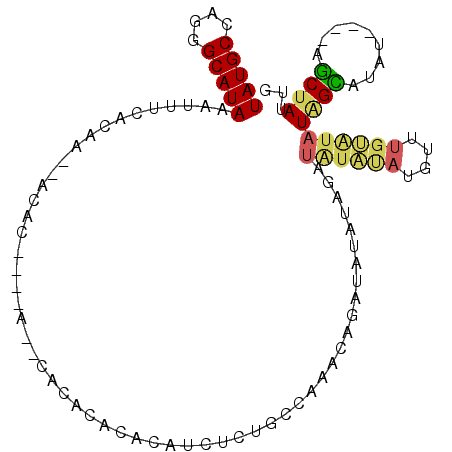
| Location | 11,301,572 – 11,301,666 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 66.00 |
| Mean single sequence MFE | -24.52 |
| Consensus MFE | -5.02 |
| Energy contribution | -7.98 |
| Covariance contribution | 2.96 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.20 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.901143 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11301572 94 - 23771897 CAAUAGCUACAUAUAUGCUAUAUACAAACAUAUAUAUCUAUAUAUCUGUAUGGCAGAGAUGUGUGUGUG--U----------UUGUAAAAUUUAUGCCCUGGCAUA ..(((((.........))))).(((((((((((((((..((...((((.....)))).)))))))))))--)----------))))).....(((((....))))) ( -28.40) >DroSec_CAF1 25710 92 - 1 CAAUAGCU----AUAUGCCAUAUACAAACAUAUACAUAUCUAUAUCUGUUUGGCAGAGAUGUGUGUGUGUGU----------CUGUGAAAUUUAUGCCCUGGCAUA .....((.----....))((((.((..(((((((((((((....((((.....)))))))))))))))))))----------.)))).....(((((....))))) ( -27.20) >DroSim_CAF1 29944 102 - 1 CAAUAGCU----AUAUGCUAUAUACAAACAUAUAUAUCUAUAUUUCUGUUUGGCAGAGAUGUGUGUGUGUGUGUCUGUGUUUUUGUGAAAUUUAUGCCCUGGCAUA ..(((((.----....)))))(((((.((((((((((.((((((((((.....)))))))))).)))))))))).)))))............(((((....))))) ( -35.30) >DroEre_CAF1 29303 82 - 1 CACUAGCU----AUAUAC-AUACUGGUACUUAUAUAUCUAU--AUCUUUUU-GCAGAUAUGU--------------GUGU--UUGUGAAAUUUAUGCCCUGGCAUA .......(----(((.((-((((.((((......)))).((--((((....-..))))))))--------------))))--.)))).....(((((....))))) ( -15.90) >DroPer_CAF1 32446 85 - 1 CAAUAGGU----AAACCCCAUGGCCAUGGAUGG-UGUGUGG--AGUGUGGAG-------UGUGUGGA-------GUGUGUGUUCGUGAAAUUUAUGCCCUGGCAUA ........----..((.(((((((((....)))-).)))))--.)).....(-------(((..((.-------(((((.(((.....))).)))))))..)))). ( -15.80) >consensus CAAUAGCU____AUAUGCCAUAUACAAACAUAUAUAUCUAUAUAUCUGUUUGGCAGAGAUGUGUGUGUG__U____GUGU__UUGUGAAAUUUAUGCCCUGGCAUA ...........................((((((((((.......((((.....)))).))))))))))........................(((((....))))) ( -5.02 = -7.98 + 2.96)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:09:37 2006