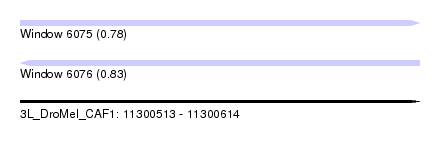
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 11,300,513 – 11,300,614 |
| Length | 101 |
| Max. P | 0.826231 |

| Location | 11,300,513 – 11,300,614 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 85.42 |
| Mean single sequence MFE | -30.57 |
| Consensus MFE | -20.46 |
| Energy contribution | -21.96 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.778885 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11300513 101 + 23771897 UUGGCUUCCAAAAAGGGAAGGG-----GGGGUUCCUAAGAACGAGGCCUAAGGUUCGAAUGGAUUGGAGGAUUUCCAAAGAAUUUCCCGGAAGAAAAGCUUAGAUU- ((..(((((.....((((((.(-----(..((((((((...((((.(....).))))......)))).))))..))......)))))))))))..)).........- ( -26.30) >DroSec_CAF1 24636 102 + 1 UUGGCUUCCAAAAAGGGAAGGG-----GGGUUUCCUAAGAACGAGGCCUAAGGUCCGAAUGGGUUGGAGGAUUUCCAACGAAUUUCCCGGAAGAAAAGCUAAGAUUC (((((((((......))))(((-----..((((........((.((((...)))))).....(((((((...)))))))))))..))).........)))))..... ( -30.50) >DroSim_CAF1 28889 100 + 1 UUGGCUACCAAAAAGGGAAGGG-----GGGU-UCCUAAGAACGAGGCCUAAGGUCCGAAUGGGUUGGAGGAUUUCCAACGAAUUUCCCGGAAGAAAAGCAAAGAUU- ...(((.((.....))...(((-----..((-((.......((.((((...)))))).....(((((((...)))))))))))..)))........))).......- ( -29.40) >DroEre_CAF1 28214 99 + 1 UUGGGUUCCUA-------AAGGGGAUGGGGGUUCCUAAGAACGAGGCCGAAGGUCCGGAUGGGUUGGAGGAUUUCCAACCCAUUUCCCGGAAGAAAACCUAAGAUC- ((((((((((.-------...))))(((((((((....))))..((.(....).)).((((((((((((...))))))))))))))))).......))))))....- ( -36.10) >consensus UUGGCUUCCAAAAAGGGAAGGG_____GGGGUUCCUAAGAACGAGGCCUAAGGUCCGAAUGGGUUGGAGGAUUUCCAACGAAUUUCCCGGAAGAAAAGCUAAGAUU_ (((((((.......((((((.......((((.((((..((.((.((((...))))))......))..)))).))))......)))))).......)))))))..... (-20.46 = -21.96 + 1.50)

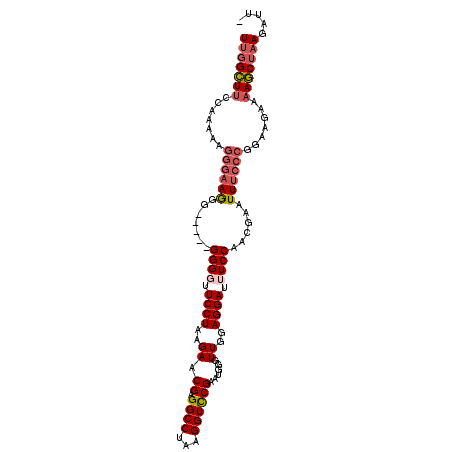

| Location | 11,300,513 – 11,300,614 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 85.42 |
| Mean single sequence MFE | -24.10 |
| Consensus MFE | -16.49 |
| Energy contribution | -16.93 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.826231 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11300513 101 - 23771897 -AAUCUAAGCUUUUCUUCCGGGAAAUUCUUUGGAAAUCCUCCAAUCCAUUCGAACCUUAGGCCUCGUUCUUAGGAACCCC-----CCCUUCCCUUUUUGGAAGCCAA -.......((((((.....(((((.....(((((.....)))))...............((....((((....))))...-----.))))))).....))))))... ( -19.70) >DroSec_CAF1 24636 102 - 1 GAAUCUUAGCUUUUCUUCCGGGAAAUUCGUUGGAAAUCCUCCAACCCAUUCGGACCUUAGGCCUCGUUCUUAGGAAACCC-----CCCUUCCCUUUUUGGAAGCCAA ((((....((((....((((((......((((((.....))))))...))))))....))))...))))...(....)..-----..(((((......))))).... ( -22.50) >DroSim_CAF1 28889 100 - 1 -AAUCUUUGCUUUUCUUCCGGGAAAUUCGUUGGAAAUCCUCCAACCCAUUCGGACCUUAGGCCUCGUUCUUAGGA-ACCC-----CCCUUCCCUUUUUGGUAGCCAA -.......(((........(((((....((((((.....))))))......((..((((((.......)))))).-.)).-----...)))))........)))... ( -20.39) >DroEre_CAF1 28214 99 - 1 -GAUCUUAGGUUUUCUUCCGGGAAAUGGGUUGGAAAUCCUCCAACCCAUCCGGACCUUCGGCCUCGUUCUUAGGAACCCCCAUCCCCUU-------UAGGAACCCAA -......(((((....(((((...((((((((((.....))))))))))))))).....))))).((((((((((.......)))....-------))))))).... ( -33.80) >consensus _AAUCUUAGCUUUUCUUCCGGGAAAUUCGUUGGAAAUCCUCCAACCCAUUCGGACCUUAGGCCUCGUUCUUAGGAAACCC_____CCCUUCCCUUUUUGGAAGCCAA ........((((((..((((((......((((((.....))))))...))))))(((.(((......))).)))........................))))))... (-16.49 = -16.93 + 0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:09:35 2006