
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 11,282,987 – 11,283,128 |
| Length | 141 |
| Max. P | 0.900685 |

| Location | 11,282,987 – 11,283,095 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 89.13 |
| Mean single sequence MFE | -21.95 |
| Consensus MFE | -19.91 |
| Energy contribution | -19.73 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.900685 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11282987 108 + 23771897 CAAGAUGACGGCACACAAGUUGACCGCAACCAAUCCGAAUUAGUUUCGGAUUAUCUCCUACACAAUUGCACAAGAUAAUUGAGGUAGCAGGGCAUAUAUAUACAUAUA ....(((.((((......)))).((((.(((((((((((.....))))))))..........(((((((....).)))))).))).)).))...........)))... ( -23.30) >DroSec_CAF1 7215 104 + 1 AAAGAUGACGGCACACAACUUGACCGCAACCAAUCCGAAUUAGUUUCGGAUUAUCUCCUACACAAUUGCACAAGAUAAUUGAGGUAGCAGGACAUAC----AAACAUA .....((.(((((.......)).)))))...((((((((.....)))))))).(((.((((.(((((((....).))))))..)))).)))......----....... ( -21.60) >DroSim_CAF1 9999 95 + 1 AAAGAUGACGGCACACAACUUGACCGCAACCAAUCCGAAUUAGUUUCGGAUUAUCUCCUACACAAUUGCACAAGAUAAUUGAGGUAGC-------------AAACAUA ....(((..((((.......)).))((.(((((((((((.....))))))))..........(((((((....).)))))).))).))-------------...))). ( -20.80) >DroEre_CAF1 7441 100 + 1 AAAGAUGACGGCACACAACUUGACCGCAACCAAUCCGAAUUAGUUUCGGAUUAUCUCUUACACAAUUGCACCAGAUAAUUGAGGUAGCAGGGCAUA--------CAUA ....(((...((.((.....)).((((.(((((((((((.....))))))))..........(((((((....).)))))).))).)).))))...--------))). ( -22.10) >consensus AAAGAUGACGGCACACAACUUGACCGCAACCAAUCCGAAUUAGUUUCGGAUUAUCUCCUACACAAUUGCACAAGAUAAUUGAGGUAGCAGGGCAUA_____AAACAUA ....(((..((((.......)).))((.(((((((((((.....))))))))..........(((((((....).)))))).))).))................))). (-19.91 = -19.73 + -0.19)

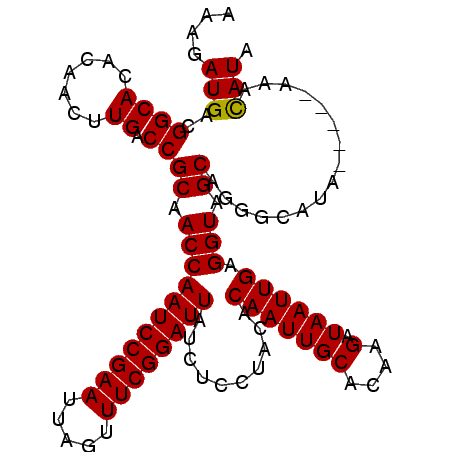

| Location | 11,282,987 – 11,283,095 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 89.13 |
| Mean single sequence MFE | -27.40 |
| Consensus MFE | -24.00 |
| Energy contribution | -24.25 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.834180 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11282987 108 - 23771897 UAUAUGUAUAUAUAUGCCCUGCUACCUCAAUUAUCUUGUGCAAUUGUGUAGGAGAUAAUCCGAAACUAAUUCGGAUUGGUUGCGGUCAACUUGUGUGCCGUCAUCUUG .....(((((((......((((.(((....(((((((.((((....)))).)))))))((((((.....))))))..))).))))......))))))).......... ( -31.00) >DroSec_CAF1 7215 104 - 1 UAUGUUU----GUAUGUCCUGCUACCUCAAUUAUCUUGUGCAAUUGUGUAGGAGAUAAUCCGAAACUAAUUCGGAUUGGUUGCGGUCAAGUUGUGUGCCGUCAUCUUU .(((...----(((..(...((((((.((((((((((.((((....)))).)))))((((((((.....))))))))))))).)))..))).)..)))...))).... ( -27.40) >DroSim_CAF1 9999 95 - 1 UAUGUUU-------------GCUACCUCAAUUAUCUUGUGCAAUUGUGUAGGAGAUAAUCCGAAACUAAUUCGGAUUGGUUGCGGUCAAGUUGUGUGCCGUCAUCUUU .......-------------((.(((....(((((((.((((....)))).)))))))((((((.....))))))..))).))(((((.....)).)))......... ( -25.10) >DroEre_CAF1 7441 100 - 1 UAUG--------UAUGCCCUGCUACCUCAAUUAUCUGGUGCAAUUGUGUAAGAGAUAAUCCGAAACUAAUUCGGAUUGGUUGCGGUCAAGUUGUGUGCCGUCAUCUUU ...(--------(((((...((((((.(((((((((..((((....))))..))))((((((((.....))))))))))))).)))..))).)))))).......... ( -26.10) >consensus UAUGUUU_____UAUGCCCUGCUACCUCAAUUAUCUUGUGCAAUUGUGUAGGAGAUAAUCCGAAACUAAUUCGGAUUGGUUGCGGUCAAGUUGUGUGCCGUCAUCUUU ....................((.(((....(((((((.((((....)))).)))))))((((((.....))))))..))).))(((((.....)).)))......... (-24.00 = -24.25 + 0.25)

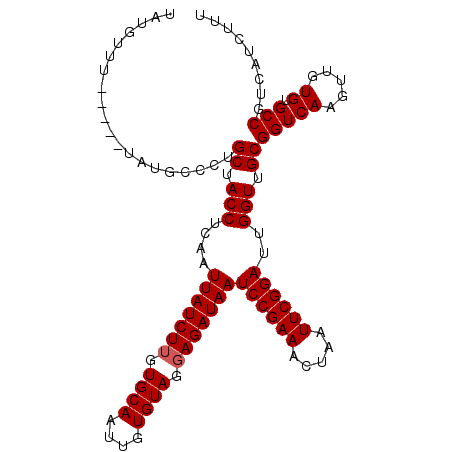

| Location | 11,283,027 – 11,283,128 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 82.17 |
| Mean single sequence MFE | -23.25 |
| Consensus MFE | -16.51 |
| Energy contribution | -18.20 |
| Covariance contribution | 1.69 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.563079 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11283027 101 - 23771897 UUGGGCCUUCCAC-----C-AAAUC-CGAGUGGUUUGAUAUAUAUGUAUAUAUAUGCCCUGCUACCUCAAUUAUCUUGUGCAAUUGUGUAGGAGAUAAUCCGAAACUA ..((((...((((-----(-.....-.).))))....(((((((....))))))))))).......((.((((((((.((((....)))).))))))))..))..... ( -25.00) >DroSec_CAF1 7255 91 - 1 UUGGGC------------CAAAAUC-CGAGUGGUUUGAUAUAUGUUU----GUAUGUCCUGCUACCUCAAUUAUCUUGUGCAAUUGUGUAGGAGAUAAUCCGAAACUA ((((..------------.......-.(((((((..(((((((....----)))))))..)))).))).((((((((.((((....)))).))))))))))))..... ( -26.90) >DroSim_CAF1 10039 90 - 1 UUGGGCCUUCCAC-----CAAAAUCACGAGUGGUUUGAUAUAUGUUU-------------GCUACCUCAAUUAUCUUGUGCAAUUGUGUAGGAGAUAAUCCGAAACUA ((((........)-----)))..((..(((((((..(((....))).-------------)))).))).((((((((.((((....)))).))))))))..))..... ( -20.00) >DroEre_CAF1 7481 99 - 1 UUGGGCCUUCCACUCCCUCCAACUC-CGAGUGGUUUGAUAUAUG--------UAUGCCCUGCUACCUCAAUUAUCUGGUGCAAUUGUGUAAGAGAUAAUCCGAAACUA ..((((...((((((..........-.))))))....(((....--------))))))).......((.(((((((..((((....))))..)))))))..))..... ( -21.10) >consensus UUGGGCCUUCCAC_____CAAAAUC_CGAGUGGUUUGAUAUAUGUUU_____UAUGCCCUGCUACCUCAAUUAUCUUGUGCAAUUGUGUAGGAGAUAAUCCGAAACUA ((((.......................(((((((....(((((........)))))....)))).))).((((((((.((((....)))).))))))))))))..... (-16.51 = -18.20 + 1.69)

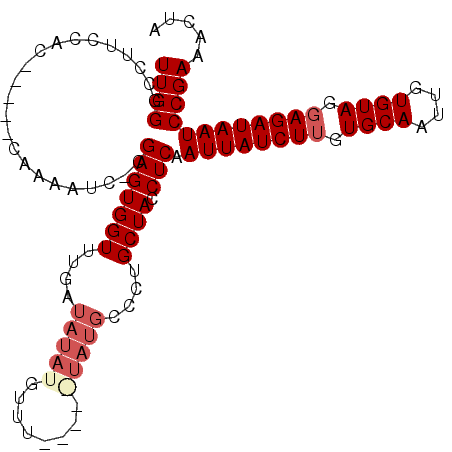
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:09:29 2006