
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 11,278,293 – 11,278,430 |
| Length | 137 |
| Max. P | 0.827365 |

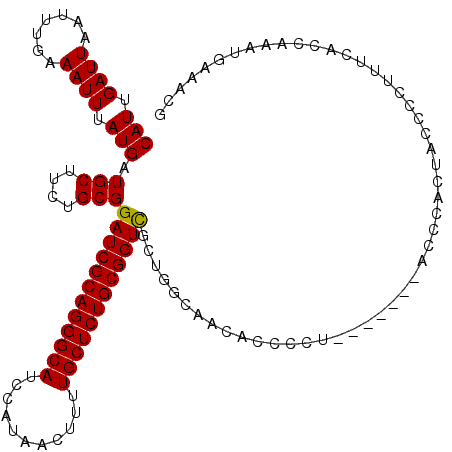
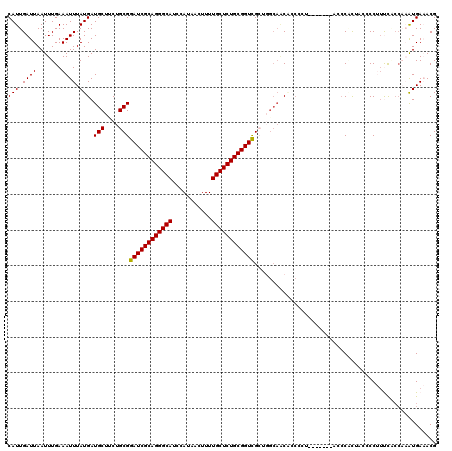
| Location | 11,278,293 – 11,278,405 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.42 |
| Mean single sequence MFE | -32.12 |
| Consensus MFE | -22.09 |
| Energy contribution | -21.87 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.827365 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11278293 112 - 23771897 CAUUGAUUAAUUUGAAAUUUAUGAUGCUUCUGCGGAUCGCAGGGCAUCCAUAACUUUUGCUCUGCGGUCGCAGACUACG--------CGCUACCCGCUACUCCUUUUCCCAAAUGAAACG .........(((((..............((((((.(((((((((((...........)))))))))))))))))....(--------((.....)))............)))))...... ( -32.40) >DroPse_CAF1 3817 110 - 1 CAUUGAUUAAUUUGAAAUUUAUGAUGCGACUGCGGAUCGCAGGGCAUCCAUAACUUUUGCUCUGCGGUUG---GGAGCACCCCUGC-----ACCCUUGCG-CCCUCCACCCCAUGA-ACG .................((((((.((.((..(((((((((((((((...........))))))))))))(---((.(((....)))-----.)))...))-)..))))...)))))-).. ( -34.10) >DroSec_CAF1 2513 108 - 1 CAUUGAUUAAUUUGAAAUUUAUGAUGCUUCUGCGGAUCGCAGGGCAUCCAUAAGUUUUGCUCUGCGGUCGCUGGCAACUCCC------------CACUACCCCUUUUGCCAAAUGAAACG ((((...........................(((.(((((((((((...........))))))))))))))((((((.....------------...........))))))))))..... ( -28.59) >DroSim_CAF1 4783 119 - 1 CAUUGAUUAAUUUGAAAUUUAUGAUGCUUCUGCGGAUCGCAGGGCAUCCAUAACUUUUGCUCUGCGGUCGCUGGCAACUCCCCUGCCCACCACCCACUACCCCU-UUGCCAAAUGAAACG .................(((((...((....(((.(((((((((((...........)))))))))))))).((((.......)))).................-..))...)))))... ( -29.60) >DroEre_CAF1 2621 116 - 1 CAUUGAUUAAUUUGAAAUUUAUGAUGCUUCUGCGGAUCGCAGGGCAUCCAUAACUUUUGCUCUGCGGUCGCUGGCACCAACCCG----GCUAACCGCUACCCCUUUCCAGAAGUGAAACG (((.((((.......)))).)))..(((((((.(((((((((((((...........))))))))))))(((((.......)))----))................))))))))...... ( -36.40) >DroPer_CAF1 3883 110 - 1 CAUUGAUUAAUUUGAAAUUUAUGAUGCGACUGCGGAUCGCAGGGCAUCCAUAACUUUUGCUCUGCGGUUG---GCAGCACCCCUGC-----ACCCUUGAG-CCCUCCACCCCAUGA-ACG .................((((((....(.((((.((((((((((((...........)))))))))))).---)))))......((-----........)-).........)))))-).. ( -31.60) >consensus CAUUGAUUAAUUUGAAAUUUAUGAUGCUUCUGCGGAUCGCAGGGCAUCCAUAACUUUUGCUCUGCGGUCGCUGGCAACACCCCU_______ACCCACUACCCCUUUCACCAAAUGAAACG (((.((((.......)))).))).(((....)))((((((((((((...........))))))))))))................................................... (-22.09 = -21.87 + -0.22)

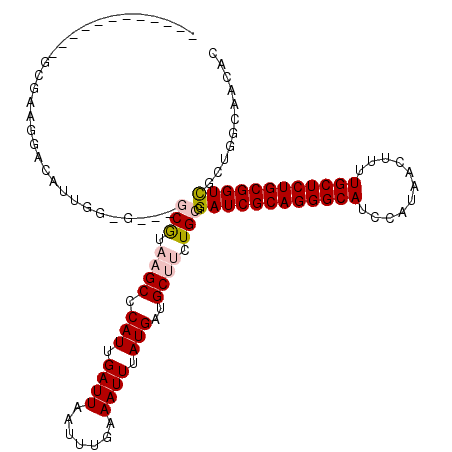
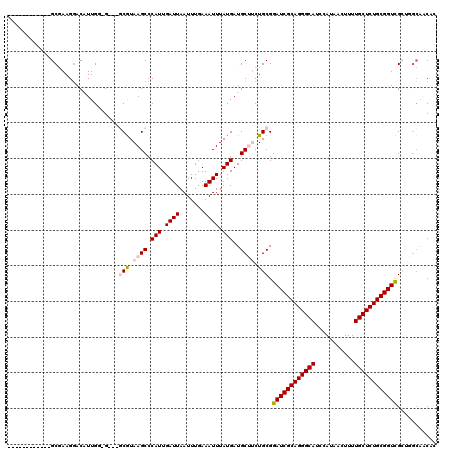
| Location | 11,278,326 – 11,278,430 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.51 |
| Mean single sequence MFE | -40.27 |
| Consensus MFE | -25.08 |
| Energy contribution | -25.63 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.98 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.728044 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11278326 104 - 23771897 ------------GCGAAGGACAUUGGGG---GCGUAAGCCCAUUGAUUAAUUUGAAAUUUAUGAUGCUUCUGCGGAUCGCAGGGCAUCCAUAACUUUUGCUCUGCGGUCGCAGACUACG- ------------.((((....((..(((---((....)))).)..))...))))..............((((((.(((((((((((...........))))))))))))))))).....- ( -40.50) >DroPse_CAF1 3850 117 - 1 GGCAGCAGGACGGCCAAGGACUUUGUUGUCCUCAUAAGCCCAUUGAUUAAUUUGAAAUUUAUGAUGCGACUGCGGAUCGCAGGGCAUCCAUAACUUUUGCUCUGCGGUUG---GGAGCAC ....((((((((((.((....)).)))))))(((((((....(..(.....)..)..))))))))))(.((.(.((((((((((((...........)))))))))))).---).))).. ( -38.60) >DroSec_CAF1 2541 104 - 1 ------------GCGAAGAACAUUGG-G---GCGUAAGCCCAUUGAUUAAUUUGAAAUUUAUGAUGCUUCUGCGGAUCGCAGGGCAUCCAUAAGUUUUGCUCUGCGGUCGCUGGCAACUC ------------.((((...((...(-(---((....))))..)).....))))..........((((...(((.(((((((((((...........)))))))))))))).)))).... ( -38.60) >DroSim_CAF1 4822 104 - 1 ------------GCGAAGGACAUUGG-G---GCGUAAGCCCAUUGAUUAAUUUGAAAUUUAUGAUGCUUCUGCGGAUCGCAGGGCAUCCAUAACUUUUGCUCUGCGGUCGCUGGCAACUC ------------.((((...((...(-(---((....))))..)).....))))..........((((...(((.(((((((((((...........)))))))))))))).)))).... ( -38.60) >DroEre_CAF1 2657 103 - 1 ------------GCGAAGGACAUUG--G---GCGUAAGCCCAUUGAUUAAUUUGAAAUUUAUGAUGCUUCUGCGGAUCGCAGGGCAUCCAUAACUUUUGCUCUGCGGUCGCUGGCACCAA ------------.....((.((.((--(---((....))))).))...................((((...(((.(((((((((((...........)))))))))))))).)))))).. ( -42.20) >DroPer_CAF1 3916 117 - 1 GGCAGCAGGACGGCCAAGGACUUUGUUGUCCUCAUAAGCCCAUUGAUUAAUUUGAAAUUUAUGAUGCGACUGCGGAUCGCAGGGCAUCCAUAACUUUUGCUCUGCGGUUG---GCAGCAC ....((((((((((.((....)).)))))))(((((((....(..(.....)..)..))))))))))(.((((.((((((((((((...........)))))))))))).---))))).. ( -43.10) >consensus ____________GCGAAGGACAUUGG_G___GCGUAAGCCCAUUGAUUAAUUUGAAAUUUAUGAUGCUUCUGCGGAUCGCAGGGCAUCCAUAACUUUUGCUCUGCGGUCGCUGGCAACAC ...............................(((.((((.(((.((((.......)))).)))..)))).))).((((((((((((...........))))))))))))........... (-25.08 = -25.63 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:09:25 2006