
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 11,267,818 – 11,267,928 |
| Length | 110 |
| Max. P | 0.938978 |

| Location | 11,267,818 – 11,267,911 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 85.61 |
| Mean single sequence MFE | -46.63 |
| Consensus MFE | -36.40 |
| Energy contribution | -35.52 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.542463 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11267818 93 - 23771897 CUUACCGCUCGCAUUGCGGCCACUGUGGCCGCCGUUGCUGCCGCUGCGGCUGCAGCAAACGCUCGAGAUCGGGAGC------GGGAUGUGGAUACCGCU ....((((((((...((((((.....))))))..(((((((.((....)).)))))))..))(((....)))))))------))...((((...)))). ( -44.80) >DroSec_CAF1 6117 93 - 1 CUUACCGCUCGUAUUGCGGCCACUGUGGCCGCCGUUGCUGCCGCUGCGGCGGCUGCAAACGCUCGAGAUCGGGAGC------GGGAUGUGGAUACCGCU ....((((((.(((..((((...((..((((((((.((....)).))))))))..))...)).))..)).).))))------))...((((...)))). ( -45.60) >DroSim_CAF1 2496 93 - 1 CUUACCGCUCGUAUUGCGGCCACUGUGGCCGCCGUUGCUGCCGCUGCGGCGGCUGCCAACGCUCGAGAUCGGGAGC------GGGAUGUGGAUACCGCU ....((((((.....((((((.....))))))((((((.(((((....))))).).))))).(((....)))))))------))...((((...)))). ( -44.60) >DroEre_CAF1 22947 93 - 1 CUUACCGCUCGCAUCGCGGCCACUGUGGCCGCCGUUGCUGCCGCUGCGGCGGCUGCAAACGCUCGAGAUCGGGAGC------GAGAUGUGGACACCGCU ......(..((((((((((((.....))))))..((((.(((((....))))).)))).(((((........))))------).))))))..)...... ( -46.10) >DroYak_CAF1 2897 93 - 1 CUUACCGCUCGCAUUGCGGCCACUGUGGCCGCCGUUGCUGCCGCAGCAGCGGUUGCAAAUGCUCGAGAUCGGGAGC------GGGAUGUGGAUACCGCU ....(((((((((((((((((.....))))((((((((((...)))))))))).)).)))))(((....)))))))------))...((((...)))). ( -47.20) >DroAna_CAF1 2527 99 - 1 CUGGCCGCCCGCUUUGCCGCUACAGUGGCGGCUGUGGCAGCUGCCGCUGCUGCCGCCAAUGCCAGGGAUCGGGAGCGGGAGCGAGAAGCGGACACCGCC ((.((..(((((((((((((....)))))(((.((((((((.......))))))))....)))........)))))))).)).))..((((...)))). ( -51.50) >consensus CUUACCGCUCGCAUUGCGGCCACUGUGGCCGCCGUUGCUGCCGCUGCGGCGGCUGCAAACGCUCGAGAUCGGGAGC______GGGAUGUGGAUACCGCU ....(((((((....((((((.....))))))((((((.(((((....))))).))).)))..))))..)))...............((((...)))). (-36.40 = -35.52 + -0.88)



| Location | 11,267,837 – 11,267,928 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 79.93 |
| Mean single sequence MFE | -42.55 |
| Consensus MFE | -30.60 |
| Energy contribution | -32.30 |
| Covariance contribution | 1.70 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.883270 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
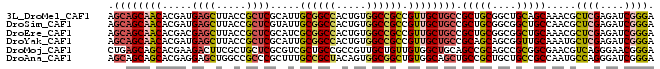
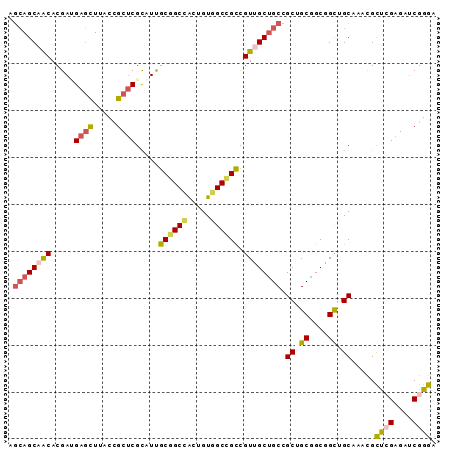
>3L_DroMel_CAF1 11267837 91 + 23771897 UCCCGAUCUCGAGCGUUUGCUGCAGCCGCAGCGGCAGCAACGGCGGCCACAGUGGCCGCAAUGCGAGCGGUAAGCUCAUCGUGUUGCUGCU ...((....)).......(((((....)))))((((((((((((((((.....)))))).....((((.....))))....)))))))))) ( -44.50) >DroSim_CAF1 2515 91 + 1 UCCCGAUCUCGAGCGUUGGCAGCCGCCGCAGCGGCAGCAACGGCGGCCACAGUGGCCGCAAUACGAGCGGUAAGCUCAUCGUGUUGCUGCU ...((....)).((..((((....))))..))((((((((((((((((.....)))))).....((((.....))))....)))))))))) ( -43.50) >DroEre_CAF1 22966 91 + 1 UCCCGAUCUCGAGCGUUUGCAGCCGCCGCAGCGGCAGCAACGGCGGCCACAGUGGCCGCGAUGCGAGCGGUAAGCUCGUCGUGUUGCUGCU ...((....))(((.......(((((....)))))((((((.((((((.....)))))).((((((((.....))))).)))))))))))) ( -44.50) >DroYak_CAF1 2916 91 + 1 UCCCGAUCUCGAGCAUUUGCAACCGCUGCUGCGGCAGCAACGGCGGCCACAGUGGCCGCAAUGCGAGCGGUAAGCUCAUCGUGUUGCUGCU ...((....))((((...((....))))))(((((((((.((((((((.....)))))).....((((.....))))..))))))))))). ( -40.70) >DroMoj_CAF1 1893 91 + 1 UCCCGUUCCCUGACGUUCGCCGCGGCUGCGGCUGCAGCCACAACAGCAACGGCGGCAGCGACGCGAGCAGCGAAGUCUUCGUGCUGCUCAG .............((((.(((((((((((....))))))....(......))))))))))....((((((((.........)))))))).. ( -37.50) >DroAna_CAF1 2552 91 + 1 UCCCGAUCCCUGGCAUUGGCGGCAGCAGCGGCAGCUGCCACAGCCGCCACUGUAGCGGCAAAGCGGGCGGCCAGCUCCUCGUGCUGCUGCU ............(((.(((((((.(((((....)))))....))))))).)))(((((((..(((((.((....)).)))))..))))))) ( -44.60) >consensus UCCCGAUCUCGAGCGUUUGCAGCCGCCGCAGCGGCAGCAACGGCGGCCACAGUGGCCGCAAUGCGAGCGGUAAGCUCAUCGUGUUGCUGCU ..................(((((....)))))((((((((((((((((.....)))))).....((((.....))))....)))))))))) (-30.60 = -32.30 + 1.70)

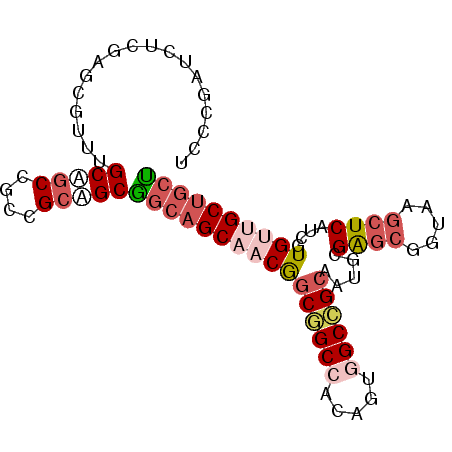
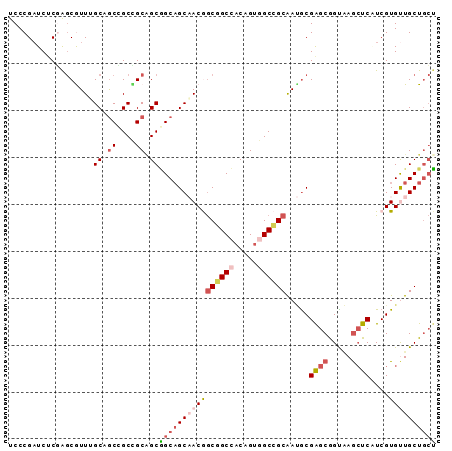
| Location | 11,267,837 – 11,267,928 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 79.93 |
| Mean single sequence MFE | -44.18 |
| Consensus MFE | -34.01 |
| Energy contribution | -34.85 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.37 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.938978 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11267837 91 - 23771897 AGCAGCAACACGAUGAGCUUACCGCUCGCAUUGCGGCCACUGUGGCCGCCGUUGCUGCCGCUGCGGCUGCAGCAAACGCUCGAGAUCGGGA .((((((((..(.(((((.....))))))...((((((.....)))))).)))))))).(((((....))))).....((((....)))). ( -45.50) >DroSim_CAF1 2515 91 - 1 AGCAGCAACACGAUGAGCUUACCGCUCGUAUUGCGGCCACUGUGGCCGCCGUUGCUGCCGCUGCGGCGGCUGCCAACGCUCGAGAUCGGGA .((((((((...((((((.....))))))...((((((.....)))))).))))))(((((....))))).)).....((((....)))). ( -43.70) >DroEre_CAF1 22966 91 - 1 AGCAGCAACACGACGAGCUUACCGCUCGCAUCGCGGCCACUGUGGCCGCCGUUGCUGCCGCUGCGGCGGCUGCAAACGCUCGAGAUCGGGA .((((((((..(.(((((.....))))))...((((((.....)))))).))))))(((((....))))).)).....((((....)))). ( -45.50) >DroYak_CAF1 2916 91 - 1 AGCAGCAACACGAUGAGCUUACCGCUCGCAUUGCGGCCACUGUGGCCGCCGUUGCUGCCGCAGCAGCGGUUGCAAAUGCUCGAGAUCGGGA .((((((((..(.(((((.....))))))...((((((.....)))))).)))))))).(((((....))))).....((((....)))). ( -43.40) >DroMoj_CAF1 1893 91 - 1 CUGAGCAGCACGAAGACUUCGCUGCUCGCGUCGCUGCCGCCGUUGCUGUUGUGGCUGCAGCCGCAGCCGCGGCGAACGUCAGGGAACGGGA ..(((((((...........))))))).((((.(((.((.((((((.((((((((....)))))))).))))))..)).))).).)))... ( -44.10) >DroAna_CAF1 2552 91 - 1 AGCAGCAGCACGAGGAGCUGGCCGCCCGCUUUGCCGCUACAGUGGCGGCUGUGGCAGCUGCCGCUGCUGCCGCCAAUGCCAGGGAUCGGGA .((((((((....(((((((.((((..(((..(((((....)))))))).))))))))).))))))))))..((....(....)....)). ( -42.90) >consensus AGCAGCAACACGAUGAGCUUACCGCUCGCAUUGCGGCCACUGUGGCCGCCGUUGCUGCCGCUGCGGCGGCUGCAAACGCUCGAGAUCGGGA .((((((((.....((((.....)))).....((((((.....)))))).)))))))).(((((....))))).....((((....)))). (-34.01 = -34.85 + 0.84)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:09:22 2006