| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 11,259,424 – 11,259,514 |
| Length | 90 |
| Max. P | 0.824029 |

| Location | 11,259,424 – 11,259,514 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 86.71 |
| Mean single sequence MFE | -17.77 |
| Consensus MFE | -12.84 |
| Energy contribution | -12.71 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.824029 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
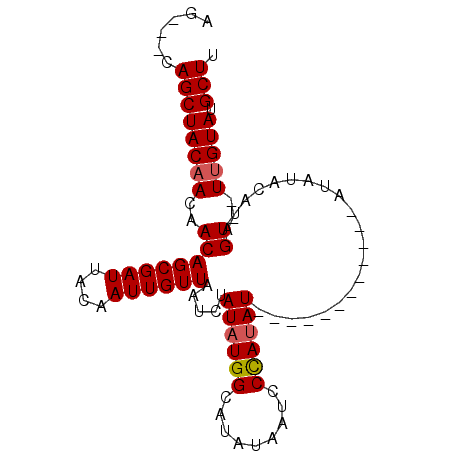
>3L_DroMel_CAF1 11259424 90 + 23771897 AAGCAUACAA--ACUAUGUAUAUAUAUACCUAUAUAUAUAGGAUUAUAUGCCAUAUAGAUUAACAAUUGUAAUCGCUGUUGUUGUAGCUG---CU ..(((((...--.(((((((((((......))))))))))).....)))))....(((..(((((((.((....)).)))))))...)))---.. ( -21.10) >DroSec_CAF1 60215 78 + 1 AAGCAUACAA--ACUAUGUAUAU------------AUAUGGGAUUAUAUGCCAUAUAGAUUAACAAUUGUAAUCGCUGUUGUUGUAGCUG---CU .(((.(((((--.....((...(------------((((((.((...)).)))))))(((((.(....)))))))).....)))))))).---.. ( -16.70) >DroSim_CAF1 61888 78 + 1 AAGCAUACCA--ACUAUGUAUAU------------AUAUGGGAUUAUAUGCCAUAUAGAUUAACAAUUGUAAUCGCUGUUGUUGUAGCUG---CU .((((((.((--((...((...(------------((((((.((...)).)))))))(((((.(....)))))))).)))).))).))).---.. ( -16.80) >DroYak_CAF1 63811 83 + 1 AAGCAUACAAAAACUAUAUUUGU------------AUAUGGGAUUAUAUGCCAAAUAGAUUAACAAUUGUAAUCGCUGUUGUUGUAGCUGCUGCU .(((.(((((.(((..(((((((------------(((((....)))))).))))))(((((.(....))))))...))).))))))))...... ( -16.50) >consensus AAGCAUACAA__ACUAUGUAUAU____________AUAUGGGAUUAUAUGCCAUAUAGAUUAACAAUUGUAAUCGCUGUUGUUGUAGCUG___CU ..(((((.((...((((((................))))))..)).)))))....(((..(((((((.((....)).)))))))...)))..... (-12.84 = -12.71 + -0.13)


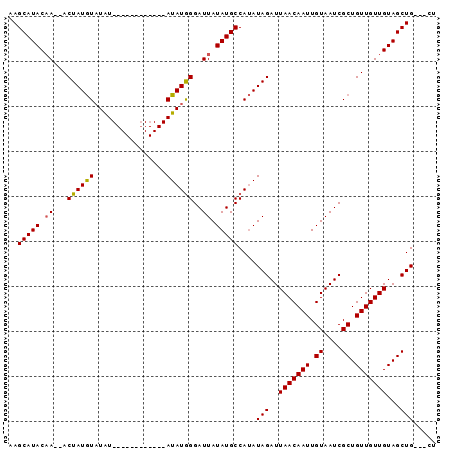
| Location | 11,259,424 – 11,259,514 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 86.71 |
| Mean single sequence MFE | -15.62 |
| Consensus MFE | -9.59 |
| Energy contribution | -9.90 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.546252 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
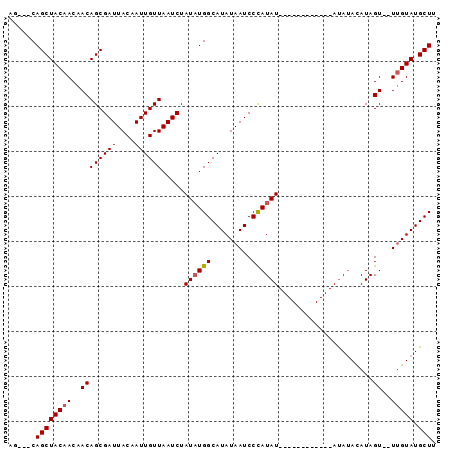
>3L_DroMel_CAF1 11259424 90 - 23771897 AG---CAGCUACAACAACAGCGAUUACAAUUGUUAAUCUAUAUGGCAUAUAAUCCUAUAUAUAUAGGUAUAUAUAUACAUAGU--UUGUAUGCUU ..---..(((........)))((((((....).))))).....(((((((((..((((.((((((......)))))).)))).--))))))))). ( -19.10) >DroSec_CAF1 60215 78 - 1 AG---CAGCUACAACAACAGCGAUUACAAUUGUUAAUCUAUAUGGCAUAUAAUCCCAUAU------------AUAUACAUAGU--UUGUAUGCUU ((---(((((........)))...(((((.(((.....(((((((.........))))))------------)...)))....--))))))))). ( -13.60) >DroSim_CAF1 61888 78 - 1 AG---CAGCUACAACAACAGCGAUUACAAUUGUUAAUCUAUAUGGCAUAUAAUCCCAUAU------------AUAUACAUAGU--UGGUAUGCUU ((---((....((((...((((((....))))))....(((((((.........))))))------------)........))--))...)))). ( -15.30) >DroYak_CAF1 63811 83 - 1 AGCAGCAGCUACAACAACAGCGAUUACAAUUGUUAAUCUAUUUGGCAUAUAAUCCCAUAU------------ACAAAUAUAGUUUUUGUAUGCUU ......((((((((.(((((((((....))))))....((((((..((((......))))------------.))))))..))).))))).))). ( -14.50) >consensus AG___CAGCUACAACAACAGCGAUUACAAUUGUUAAUCUAUAUGGCAUAUAAUCCCAUAU____________AUAUACAUAGU__UUGUAUGCUU ......((((((((..((((((((....)))))).....((((((.........)))))).....................))..))))).))). ( -9.59 = -9.90 + 0.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:09:18 2006