
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 11,259,066 – 11,259,211 |
| Length | 145 |
| Max. P | 0.999945 |

| Location | 11,259,066 – 11,259,159 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.35 |
| Mean single sequence MFE | -17.78 |
| Consensus MFE | -3.42 |
| Energy contribution | -4.62 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.19 |
| SVM decision value | 2.81 |
| SVM RNA-class probability | 0.997153 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11259066 93 + 23771897 AAGAAAACCAACAAUACUUGUAAAUAACAUUAUGGCCCAUACAGCAGCAAC---------------------------AAUAGUUGCUACAAUAACAGCAGCAACAUUGUAGCUAAAGUC ...............((((...............((.......))(((.((---------------------------(((.((((((.(.......).))))))))))).))).)))). ( -15.80) >DroSec_CAF1 59916 93 + 1 AAGAAAACCAACAAUACUUGUAAAUAACAUUAUGGCCCAUACAGCAGCAAC---------------------------AAUAGUUGCUACAAUAACAGCAGCAACAUUGUAGCUACAGUC .................(((((............((.......)).((.((---------------------------(((.((((((.(.......).))))))))))).))))))).. ( -15.90) >DroSim_CAF1 61586 93 + 1 AAGAAAACCAACAAUACUUGUAAAUAACAUUAUGGCCCAUACAGCAGCAAC---------------------------AAUAGUUGCUACAAUAACAGCAGCAACAUUGUUGCUACAGUC .................(((((............((.......)).(((((---------------------------(((.((((((.(.......).))))))))))))))))))).. ( -20.00) >DroYak_CAF1 63485 118 + 1 AUGAAAACCAACAACACUUUUAAAUAACAUUAUGGCC-AAACAGCAGCAACAAUAGUUGCUACAAUAACAGCAGAUACAAUAGUUGCUACCAUAACAGCAGCAACAUUGUGUCUA-AGUA ...............((((.............((((.-.....))((((((....)))))).))........(((((((((.((((((.(.......).))))))))))))))))-))). ( -25.30) >DroAna_CAF1 60037 92 + 1 -GCAACACCAGCAGUAUUUGUAAAUAACAGUAUUGCCAGUGCAACAACAGC---------------------------AACAAAAACCAUGAAGAAAACAACAAAUUUGAAGCCCAAGUU -....(((..((((((((.((.....))))))))))..)))..........---------------------------.........................((((((.....)))))) ( -11.90) >consensus AAGAAAACCAACAAUACUUGUAAAUAACAUUAUGGCCCAUACAGCAGCAAC___________________________AAUAGUUGCUACAAUAACAGCAGCAACAUUGUAGCUAAAGUC ..................................................................................((((((........)))))).................. ( -3.42 = -4.62 + 1.20)

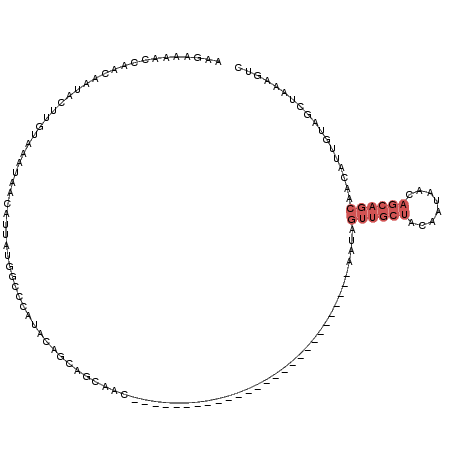
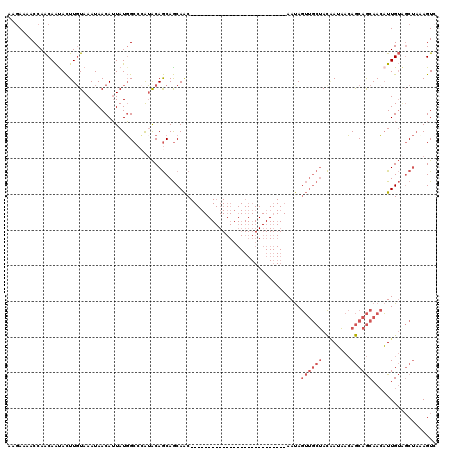
| Location | 11,259,066 – 11,259,159 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.35 |
| Mean single sequence MFE | -28.84 |
| Consensus MFE | -12.24 |
| Energy contribution | -12.44 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.31 |
| Mean z-score | -3.73 |
| Structure conservation index | 0.42 |
| SVM decision value | 4.74 |
| SVM RNA-class probability | 0.999945 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11259066 93 - 23771897 GACUUUAGCUACAAUGUUGCUGCUGUUAUUGUAGCAACUAUU---------------------------GUUGCUGCUGUAUGGGCCAUAAUGUUAUUUACAAGUAUUGUUGGUUUUCUU .((..((((.(((((((((((((.......)))))))).)))---------------------------)).))))..))..(((((((((((((.......))))))).)))))).... ( -26.40) >DroSec_CAF1 59916 93 - 1 GACUGUAGCUACAAUGUUGCUGCUGUUAUUGUAGCAACUAUU---------------------------GUUGCUGCUGUAUGGGCCAUAAUGUUAUUUACAAGUAUUGUUGGUUUUCUU .((.(((((.(((((((((((((.......)))))))).)))---------------------------)).))))).))..(((((((((((((.......))))))).)))))).... ( -29.40) >DroSim_CAF1 61586 93 - 1 GACUGUAGCAACAAUGUUGCUGCUGUUAUUGUAGCAACUAUU---------------------------GUUGCUGCUGUAUGGGCCAUAAUGUUAUUUACAAGUAUUGUUGGUUUUCUU .((.(((((((((((((((((((.......)))))))).)))---------------------------)))))))).))..(((((((((((((.......))))))).)))))).... ( -33.50) >DroYak_CAF1 63485 118 - 1 UACU-UAGACACAAUGUUGCUGCUGUUAUGGUAGCAACUAUUGUAUCUGCUGUUAUUGUAGCAACUAUUGUUGCUGCUGUUU-GGCCAUAAUGUUAUUUAAAAGUGUUGUUGGUUUUCAU ....-((((.((((((((((((((.....))))))))).))))).))))........((((((((....))))))))((...-((((((((..((.......))..))).)))))..)). ( -37.70) >DroAna_CAF1 60037 92 - 1 AACUUGGGCUUCAAAUUUGUUGUUUUCUUCAUGGUUUUUGUU---------------------------GCUGUUGUUGCACUGGCAAUACUGUUAUUUACAAAUACUGCUGGUGUUGC- ......(((..((((.....((.......)).....))))..---------------------------)))...((.((((..(((.((.(((.....)))..)).)))..)))).))- ( -17.20) >consensus GACUGUAGCUACAAUGUUGCUGCUGUUAUUGUAGCAACUAUU___________________________GUUGCUGCUGUAUGGGCCAUAAUGUUAUUUACAAGUAUUGUUGGUUUUCUU ...........(((((.(((((((..(((.((((((((...............................)))))))).)))..))).....(((.....))))))).)))))........ (-12.24 = -12.44 + 0.20)

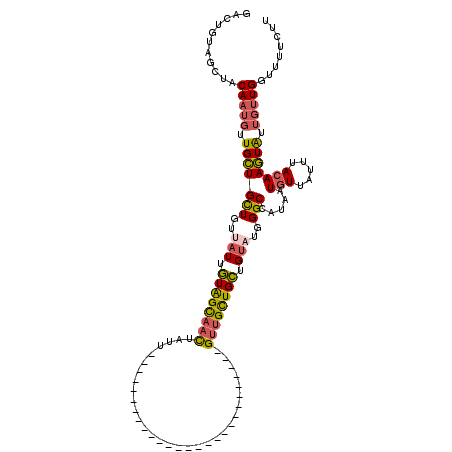

| Location | 11,259,106 – 11,259,196 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 70.10 |
| Mean single sequence MFE | -23.71 |
| Consensus MFE | -10.20 |
| Energy contribution | -11.20 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.43 |
| SVM decision value | 3.54 |
| SVM RNA-class probability | 0.999365 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11259106 90 + 23771897 ACAGCAGCAAC---------------------------AAUAGUUGCUACAAUAACAGCAGCAACAUUGUAGCUAAAGUCGUAGCCCUCUUUUGGCAGUGUCUA-AACAAUGAGAA--AA .....(((.((---------------------------(((.((((((.(.......).))))))))))).)))..((.((..(((.......)))..)).)).-...........--.. ( -20.40) >DroSec_CAF1 59956 90 + 1 ACAGCAGCAAC---------------------------AAUAGUUGCUACAAUAACAGCAGCAACAUUGUAGCUACAGUCGUAGCCCUCAUUUGGCAGUGUCUA-AACACUGAAAA--AA .........((---------------------------(((.((((((.(.......).))))))))))).(((((....))))).....(((..((((((...-.))))))..))--). ( -24.20) >DroSim_CAF1 61626 92 + 1 ACAGCAGCAAC---------------------------AAUAGUUGCUACAAUAACAGCAGCAACAUUGUUGCUACAGUCGUAGCCCUCUUUUGGCAGUGUCUA-AACACUGAAAAAAAA ...(.((((((---------------------------(((.((((((.(.......).))))))))))))))).).............((((..((((((...-.))))))..)))).. ( -27.80) >DroYak_CAF1 63524 116 + 1 ACAGCAGCAACAAUAGUUGCUACAAUAACAGCAGAUACAAUAGUUGCUACCAUAACAGCAGCAACAUUGUGUCUA-AGUAGUAGCCCUCUUUUGGCAGUGUCUG-UACACUGAGAA--AA ...((((((((....))))))......((.(((((((((((.((((((.(.......).))))))))))))))).-.)).)).))....((((..((((((...-.)))))).)))--). ( -33.20) >DroAna_CAF1 60076 91 + 1 GCAACAACAGC---------------------------AACAAAAACCAUGAAGAAAACAACAAAUUUGAAGCCCAAGUUGUUGUCCAAGUUUGGCAUUAGUUAGGGUACUGAGAA--AA ((.......))---------------------------........(((........(((((((.((((.....))))))))))).......)))..(((((......)))))...--.. ( -12.96) >consensus ACAGCAGCAAC___________________________AAUAGUUGCUACAAUAACAGCAGCAACAUUGUAGCUAAAGUCGUAGCCCUCUUUUGGCAGUGUCUA_AACACUGAGAA__AA .............................................(((((.((...(((.(((....))).)))...)).)))))((......))((((((.....))))))........ (-10.20 = -11.20 + 1.00)

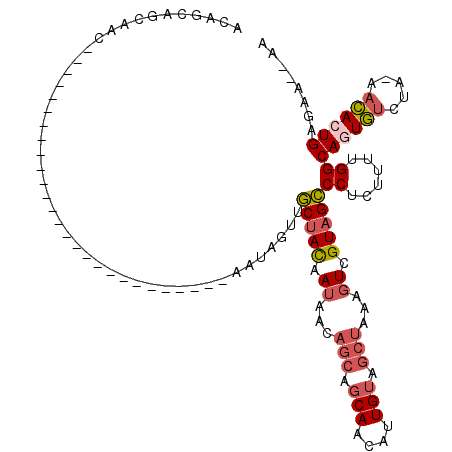
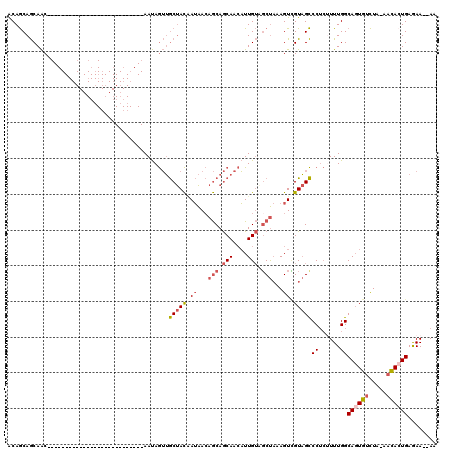
| Location | 11,259,106 – 11,259,196 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 70.10 |
| Mean single sequence MFE | -29.84 |
| Consensus MFE | -13.84 |
| Energy contribution | -13.92 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.35 |
| Mean z-score | -3.44 |
| Structure conservation index | 0.46 |
| SVM decision value | 4.43 |
| SVM RNA-class probability | 0.999895 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11259106 90 - 23771897 UU--UUCUCAUUGUU-UAGACACUGCCAAAAGAGGGCUACGACUUUAGCUACAAUGUUGCUGCUGUUAUUGUAGCAACUAUU---------------------------GUUGCUGCUGU ..--...........-........(((.......)))....((..((((.(((((((((((((.......)))))))).)))---------------------------)).))))..)) ( -24.90) >DroSec_CAF1 59956 90 - 1 UU--UUUUCAGUGUU-UAGACACUGCCAAAUGAGGGCUACGACUGUAGCUACAAUGUUGCUGCUGUUAUUGUAGCAACUAUU---------------------------GUUGCUGCUGU ..--.....(((((.-...)))))(((.......)))....((.(((((.(((((((((((((.......)))))))).)))---------------------------)).))))).)) ( -31.90) >DroSim_CAF1 61626 92 - 1 UUUUUUUUCAGUGUU-UAGACACUGCCAAAAGAGGGCUACGACUGUAGCAACAAUGUUGCUGCUGUUAUUGUAGCAACUAUU---------------------------GUUGCUGCUGU ((((((..((((((.-...))))))..))))))........((.(((((((((((((((((((.......)))))))).)))---------------------------)))))))).)) ( -37.20) >DroYak_CAF1 63524 116 - 1 UU--UUCUCAGUGUA-CAGACACUGCCAAAAGAGGGCUACUACU-UAGACACAAUGUUGCUGCUGUUAUGGUAGCAACUAUUGUAUCUGCUGUUAUUGUAGCAACUAUUGUUGCUGCUGU ((--((..((((((.-...))))))..))))((.(((.......-.(((.((((((((((((((.....))))))))).))))).)))))).))...((((((((....))))))))... ( -39.90) >DroAna_CAF1 60076 91 - 1 UU--UUCUCAGUACCCUAACUAAUGCCAAACUUGGACAACAACUUGGGCUUCAAAUUUGUUGUUUUCUUCAUGGUUUUUGUU---------------------------GCUGUUGUUGC ..--....(((((.....((....((((.....(((((((((.((........)).)))))))))......))))....)))---------------------------))))....... ( -15.30) >consensus UU__UUCUCAGUGUU_UAGACACUGCCAAAAGAGGGCUACGACUGUAGCUACAAUGUUGCUGCUGUUAUUGUAGCAACUAUU___________________________GUUGCUGCUGU ........((((((.....))))))......((.(((..((((............))))..))).))...((((((((...............................))))))))... (-13.84 = -13.92 + 0.08)

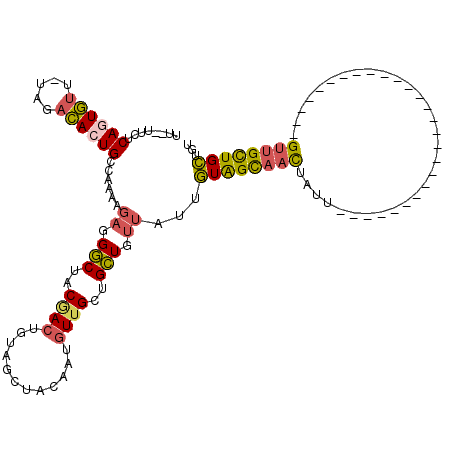
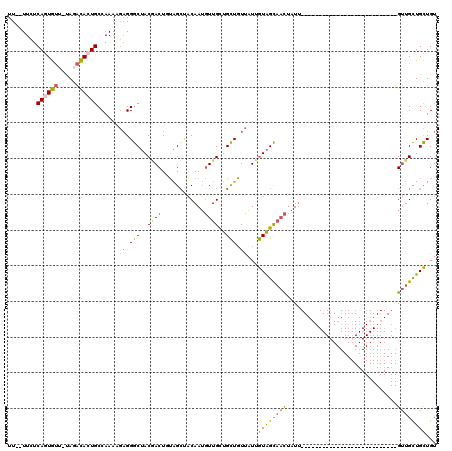
| Location | 11,259,119 – 11,259,211 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 86.20 |
| Mean single sequence MFE | -21.95 |
| Consensus MFE | -15.38 |
| Energy contribution | -16.88 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933996 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11259119 92 + 23771897 UAGUUGCUACAAUAACAGCAGCAACAUUGUAGCUAAAGUCGUAGCCCUCUUUUGGCAGUGUCUAAACAAUGAGAA--AAAUGUAUCCAAAAACG ..((((((........))))))..((((((......((.((..(((.......)))..)).))..))))))....--................. ( -16.80) >DroSec_CAF1 59969 92 + 1 UAGUUGCUACAAUAACAGCAGCAACAUUGUAGCUACAGUCGUAGCCCUCAUUUGGCAGUGUCUAAACACUGAAAA--AAAUGUAUCCAAAAAUA ..((((((........)))))).(((((...(((((....))))).....(((..((((((....))))))..))--))))))........... ( -23.90) >DroSim_CAF1 61639 94 + 1 UAGUUGCUACAAUAACAGCAGCAACAUUGUUGCUACAGUCGUAGCCCUCUUUUGGCAGUGUCUAAACACUGAAAAAAAAAUAUACCCAAAAAUA .....(((((.((...(((((((....)))))))...)).)))))....((((..((((((....))))))..))))................. ( -22.90) >DroYak_CAF1 63564 86 + 1 UAGUUGCUACCAUAACAGCAGCAACAUUGUGUCUA-AGUAGUAGCCCUCUUUUGGCAGUGUCUGUACACUGAGAA--AACUGUGGCUAA----- ..((((((........)))))).............-.....((((((..((((..((((((....))))))..))--))..).))))).----- ( -24.20) >consensus UAGUUGCUACAAUAACAGCAGCAACAUUGUAGCUACAGUCGUAGCCCUCUUUUGGCAGUGUCUAAACACUGAAAA__AAAUGUAUCCAAAAAUA ..((((((........)))))).(((((...(((((....)))))((......))((((((....)))))).......)))))........... (-15.38 = -16.88 + 1.50)

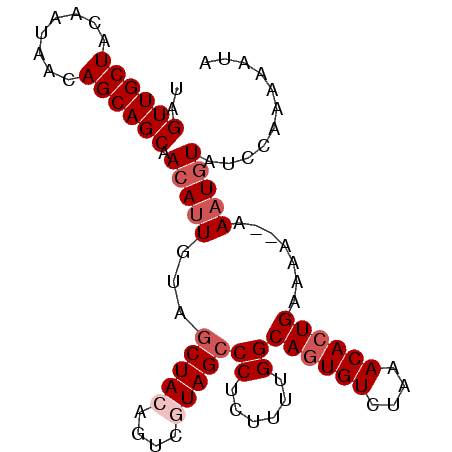
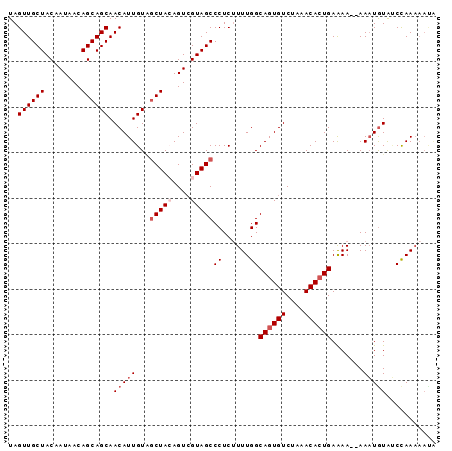
| Location | 11,259,119 – 11,259,211 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 86.20 |
| Mean single sequence MFE | -25.95 |
| Consensus MFE | -19.30 |
| Energy contribution | -20.30 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.975597 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11259119 92 - 23771897 CGUUUUUGGAUACAUUU--UUCUCAUUGUUUAGACACUGCCAAAAGAGGGCUACGACUUUAGCUACAAUGUUGCUGCUGUUAUUGUAGCAACUA ..(((((((.....(((--..(.....)...))).....)))))))..(((((......))))).....((((((((.......)))))))).. ( -19.60) >DroSec_CAF1 59969 92 - 1 UAUUUUUGGAUACAUUU--UUUUCAGUGUUUAGACACUGCCAAAUGAGGGCUACGACUGUAGCUACAAUGUUGCUGCUGUUAUUGUAGCAACUA .....(((....(((((--....((((((....))))))..)))))..((((((....)))))).))).((((((((.......)))))))).. ( -27.70) >DroSim_CAF1 61639 94 - 1 UAUUUUUGGGUAUAUUUUUUUUUCAGUGUUUAGACACUGCCAAAAGAGGGCUACGACUGUAGCAACAAUGUUGCUGCUGUUAUUGUAGCAACUA ........(((...(((((((..((((((....))))))..))))))).((((((((.((((((((...)))))))).)))...))))).))). ( -28.90) >DroYak_CAF1 63564 86 - 1 -----UUAGCCACAGUU--UUCUCAGUGUACAGACACUGCCAAAAGAGGGCUACUACU-UAGACACAAUGUUGCUGCUGUUAUGGUAGCAACUA -----.(((((.(..((--((..((((((....))))))..))))..)))))).....-..........(((((((((.....))))))))).. ( -27.60) >consensus UAUUUUUGGAUACAUUU__UUCUCAGUGUUUAGACACUGCCAAAAGAGGGCUACGACUGUAGCUACAAUGUUGCUGCUGUUAUUGUAGCAACUA .......................((((((....))))))((......))(((((....)))))......((((((((.......)))))))).. (-19.30 = -20.30 + 1.00)

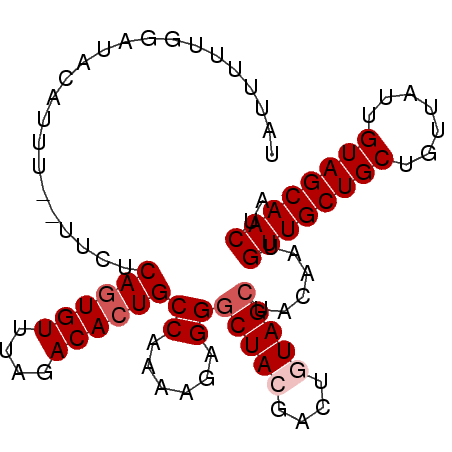
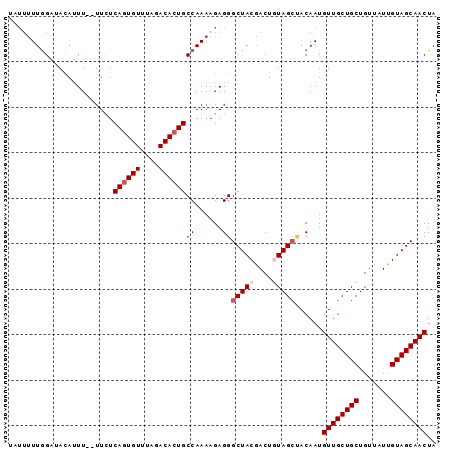
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:09:16 2006