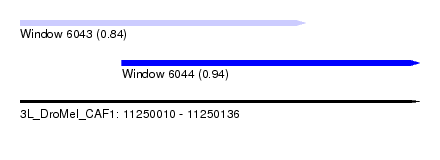
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 11,250,010 – 11,250,136 |
| Length | 126 |
| Max. P | 0.944142 |

| Location | 11,250,010 – 11,250,100 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 76.09 |
| Mean single sequence MFE | -23.26 |
| Consensus MFE | -15.81 |
| Energy contribution | -17.87 |
| Covariance contribution | 2.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.840779 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
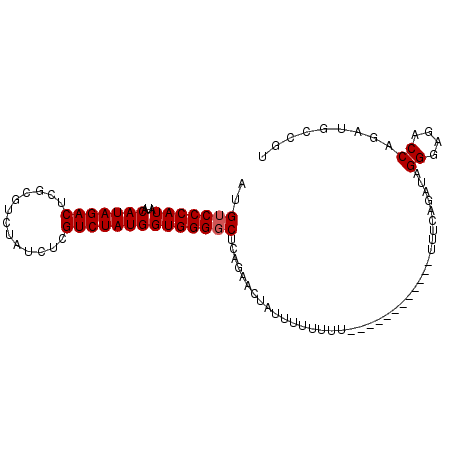
>3L_DroMel_CAF1 11250010 90 + 23771897 UCACUUGCAUUGGGGCGAUAAGUCGCUAUAUC---------AAGACCCAUAUACAUAGACAUAGA-------UGUCUAUGGUGGGGCUCAGAACUAUUUUUUUUCA ...((((.((...(((((....)))))..)))---------))).(((((...((((((((....-------)))))))))))))..................... ( -26.00) >DroSec_CAF1 51828 94 + 1 CCACUUGCAUUGGGGCGAUAAGUCGCUAUACC---------AUGUCCCAUAUACAUAGACUCGCGUCUAUCUCGUCUAUGGUGGGGCUCAGAACAAUUUU-UUU-- ..........((((((((....)))))...))---------).(((((((...(((((((.............)))))))))))))).............-...-- ( -24.72) >DroSim_CAF1 53561 95 + 1 CCACUUGCAUUGGGGCGAUAAGUCGCUAUACC---------AUGUCCCAUAUACAUAGACUCGCGUCUAUCUCGUCUAUGGUGGGGCUCAGAACUAUUUUUUUU-- ..........((((((((....)))))...))---------).(((((((...(((((((.............)))))))))))))).................-- ( -24.72) >DroYak_CAF1 55148 88 + 1 CCACUUGCAUUGGGGCGAUAAGUCGCUAUAGUACACUAUAUAUGUCCCACAUAUA---------------CUCGUCUAU-AUGAGGCUCAAAACUAUUUUUUUU-- ......((..((((((((....))))(((((....))))).....))))......---------------(((((....-))))))).................-- ( -17.60) >consensus CCACUUGCAUUGGGGCGAUAAGUCGCUAUACC_________AUGUCCCAUAUACAUAGACUCGCG_____CUCGUCUAUGGUGGGGCUCAGAACUAUUUUUUUU__ .........(((((((((....)))))................(((((((...(((((((.............))))))))))))))))))............... (-15.81 = -17.87 + 2.06)


| Location | 11,250,042 – 11,250,136 |
|---|---|
| Length | 94 |
| Sequences | 3 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 80.07 |
| Mean single sequence MFE | -22.78 |
| Consensus MFE | -20.95 |
| Energy contribution | -21.29 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.944142 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11250042 94 + 23771897 AAGACCCAUAUACAUAGACAUAGA-------UGUCUAUGGUGGGGCUCAGAACUAUUUUUUUUCAUUUUUUUUUGUUUCAGAUAGGGAGACCAGAUGCCGU .....((((...((((((((....-------))))))))))))(((((.....(((((.....((........))....)))))((....)).)).))).. ( -23.50) >DroSec_CAF1 51860 88 + 1 AUGUCCCAUAUACAUAGACUCGCGUCUAUCUCGUCUAUGGUGGGGCUCAGAACAAUUUU-UUU------------UUUCAGAUAGGGAGACCAGAUGCCGU ..(((((((...(((((((.............)))))))))))))).............-...------------.........((....))......... ( -22.42) >DroSim_CAF1 53593 89 + 1 AUGUCCCAUAUACAUAGACUCGCGUCUAUCUCGUCUAUGGUGGGGCUCAGAACUAUUUUUUUU------------UUUCAGAUAGGGAGACCAGAUGCCGU ..(((((((...(((((((.............)))))))))))))).................------------.........((....))......... ( -22.42) >consensus AUGUCCCAUAUACAUAGACUCGCGUCUAUCUCGUCUAUGGUGGGGCUCAGAACUAUUUUUUUU____________UUUCAGAUAGGGAGACCAGAUGCCGU ..(((((((...(((((((.............))))))))))))))......................................((....))......... (-20.95 = -21.29 + 0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:09:05 2006