
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 11,222,913 – 11,223,025 |
| Length | 112 |
| Max. P | 0.999978 |

| Location | 11,222,913 – 11,223,025 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 74.92 |
| Mean single sequence MFE | -28.57 |
| Consensus MFE | -17.00 |
| Energy contribution | -17.33 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -4.33 |
| Structure conservation index | 0.60 |
| SVM decision value | 4.55 |
| SVM RNA-class probability | 0.999919 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11222913 112 + 23771897 CAUUUCCAUUUGGCUGUCUGUCAUAAAUUACUUGCCAAAUUCCUUGGCAUUAACUUUGGCCGACAGCCCGGAAACAUAACUGUGUACAUACUUA---CAUAUGUAUGUAUGUAUA ..(((((....(((((((.((((....(((..((((((.....)))))).)))...)))).))))))).)))))........(((((((((.((---(....))).))))))))) ( -39.30) >DroSim_CAF1 31879 92 + 1 CAUUUCCAUUUGGCU----GUCAUAAAUUACUUGCCAAAUUCCUUGGCAUUAACUUUGGCCGACAGCCCGGAAACAUAUCUGUGUAC-------------------AUACAUAUA ..(((((....((((----(((.((((.....((((((.....)))))).....))))...))))))).)))))......(((((..-------------------..))))).. ( -25.80) >DroEre_CAF1 33052 92 + 1 CAUUUCCAUUUGGCU----GUCAUAAAUUACUUGGCAAAUUCCUUGGCAUUAACUUUGGCCGACAGCCCGGAAAC-------------------AUAUAUCUGUAUAUCUGUAUA ...((((....((((----(((...........((......))..(((..........)))))))))).))))((-------------------(.((((....)))).)))... ( -20.60) >consensus CAUUUCCAUUUGGCU____GUCAUAAAUUACUUGCCAAAUUCCUUGGCAUUAACUUUGGCCGACAGCCCGGAAACAUA_CUGUGUAC___________AU_UGUAUAUAUGUAUA ..(((((....((((....((((....(((..((((((.....)))))).)))...))))....)))).)))))......................................... (-17.00 = -17.33 + 0.33)

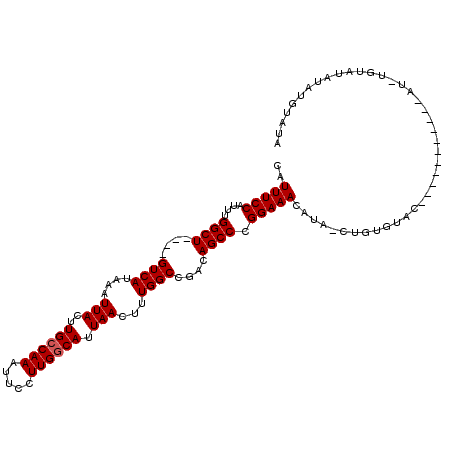

| Location | 11,222,913 – 11,223,025 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 74.92 |
| Mean single sequence MFE | -30.43 |
| Consensus MFE | -19.77 |
| Energy contribution | -20.10 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.75 |
| Structure conservation index | 0.65 |
| SVM decision value | 5.20 |
| SVM RNA-class probability | 0.999978 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11222913 112 - 23771897 UAUACAUACAUACAUAUG---UAAGUAUGUACACAGUUAUGUUUCCGGGCUGUCGGCCAAAGUUAAUGCCAAGGAAUUUGGCAAGUAAUUUAUGACAGACAGCCAAAUGGAAAUG ..(((((((.(((....)---)).))))))).........((((((.(((((((.(.((((((((.(((((((...)))))))..)))))).)).).)))))))....)))))). ( -38.30) >DroSim_CAF1 31879 92 - 1 UAUAUGUAU-------------------GUACACAGAUAUGUUUCCGGGCUGUCGGCCAAAGUUAAUGCCAAGGAAUUUGGCAAGUAAUUUAUGAC----AGCCAAAUGGAAAUG ...((((.(-------------------(....)).))))((((((.((((((((....((((((.(((((((...)))))))..)))))).))))----))))....)))))). ( -29.20) >DroEre_CAF1 33052 92 - 1 UAUACAGAUAUACAGAUAUAU-------------------GUUUCCGGGCUGUCGGCCAAAGUUAAUGCCAAGGAAUUUGCCAAGUAAUUUAUGAC----AGCCAAAUGGAAAUG .....................-------------------((((((.((((((((((..........)))..((......))...........)))----))))....)))))). ( -23.80) >consensus UAUACAUAUAUACA_AU___________GUACACAG_UAUGUUUCCGGGCUGUCGGCCAAAGUUAAUGCCAAGGAAUUUGGCAAGUAAUUUAUGAC____AGCCAAAUGGAAAUG ........................................((((((.((((((((....((((((.(((((((...)))))))..)))))).))))....))))....)))))). (-19.77 = -20.10 + 0.33)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:08:55 2006