
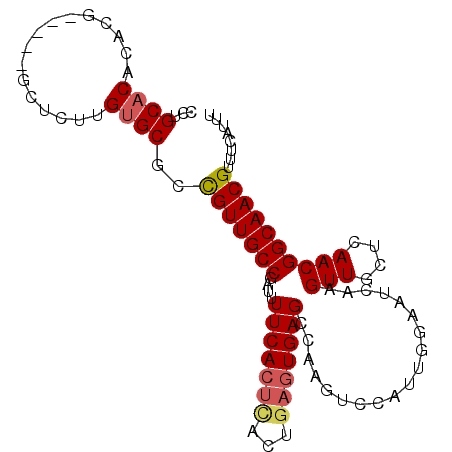
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 11,220,317 – 11,220,413 |
| Length | 96 |
| Max. P | 0.927636 |

| Location | 11,220,317 – 11,220,413 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 78.76 |
| Mean single sequence MFE | -32.83 |
| Consensus MFE | -19.81 |
| Energy contribution | -20.93 |
| Covariance contribution | 1.13 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.909420 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11220317 96 + 23771897 AAAUGAAACGUUGCCGUUGAGCAACUUGAUUCCAAUGGACUUGGCUCACUCAGUGAGUGAAAAACGGCAACGGCGCACGAGAGC-----CGUGUGUGCAGG ........((((((((((............(((...))).((.((((((...)))))).)).))))))))))(((((((.....-----)))))))..... ( -34.60) >DroVir_CAF1 31378 81 + 1 GAGCGCGACGUUGCCGUUGUACAACUCGUUUGCAGGCGGCUCAGCUCACAAUGUGAGUGAAAAACGGCAACAAGGC--------------------GCAGG ..((((...(((((((((.....((((..(((..(((......)))..)))...))))....)))))))))...))--------------------))... ( -32.70) >DroSec_CAF1 29081 96 + 1 AAAUGAAACGUUGCCGUUGAGCAACUUGAUUCCAAUGGACUUGGCUCACUCAGUGAGUGAAAAACGGCAACGGCGCACAAGAGC-----CGUGUGUGCAGG ........((((((((((............(((...))).((.((((((...)))))).)).))))))))))(((((((.....-----..)))))))... ( -34.80) >DroSim_CAF1 29302 96 + 1 AAAUGAAACGUUGCCGUUGAGCAACUUGAUUCCAAUGGACUUGGCUCACUCAGUGAGUGAAAAUCGGCAACGGCGCACAAGAGC-----CGUGUGUGCAGG ........(((((((((..(((...(((....)))..).))..))((((((...)))))).....)))))))(((((((.....-----..)))))))... ( -34.80) >DroYak_CAF1 31277 96 + 1 AAAUGAAACGUUGCCGUUGAGCAACUUGAUUCCAAUGGACGCGGCUCACUCAGUGAGUGAAAAUUGGCAACGGCGCACAAGAGC-----CGUGUGUGCAGG ........(((((((((((..(...(((....)))..)...))))((((((...)))))).....)))))))(((((((.....-----..)))))))... ( -31.70) >DroAna_CAF1 30482 99 + 1 -GAUGAAACGUUGCCGUUGAGCAACUGGCUUCCAGGCCGCUUUUCUCACUCAGUAACUGAAAAUCGGCAACGCUGCCCCU-AGCCCCAUCGUGUGUGCAGG -((((....(((((((.((((....(((((....))))).....)))).((((...))))....)))))))((((....)-)))..))))........... ( -28.40) >consensus AAAUGAAACGUUGCCGUUGAGCAACUUGAUUCCAAUGGACUUGGCUCACUCAGUGAGUGAAAAACGGCAACGGCGCACAAGAGC_____CGUGUGUGCAGG ........((((((((((....)))..................((((((...)))))).......)))))))..(((((...((......)).)))))... (-19.81 = -20.93 + 1.13)

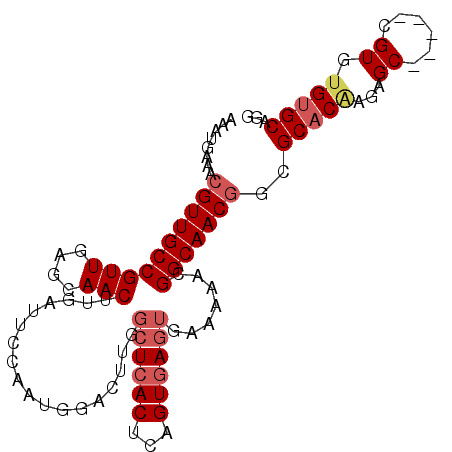
| Location | 11,220,317 – 11,220,413 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 78.76 |
| Mean single sequence MFE | -30.73 |
| Consensus MFE | -19.14 |
| Energy contribution | -19.64 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.927636 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11220317 96 - 23771897 CCUGCACACACG-----GCUCUCGUGCGCCGUUGCCGUUUUUCACUCACUGAGUGAGCCAAGUCCAUUGGAAUCAAGUUGCUCAACGGCAACGUUUCAUUU ...((((.....-----......))))..((((((((((.(((((((...)))))))((((.....)))).............))))))))))........ ( -29.70) >DroVir_CAF1 31378 81 - 1 CCUGC--------------------GCCUUGUUGCCGUUUUUCACUCACAUUGUGAGCUGAGCCGCCUGCAAACGAGUUGUACAACGGCAACGUCGCGCUC ...((--------------------((..((((((((((...(((((...((((..((......))..))))..))).))...))))))))))..)))).. ( -32.60) >DroSec_CAF1 29081 96 - 1 CCUGCACACACG-----GCUCUUGUGCGCCGUUGCCGUUUUUCACUCACUGAGUGAGCCAAGUCCAUUGGAAUCAAGUUGCUCAACGGCAACGUUUCAUUU ...(((((....-----.....)))))..((((((((((.(((((((...)))))))((((.....)))).............))))))))))........ ( -30.70) >DroSim_CAF1 29302 96 - 1 CCUGCACACACG-----GCUCUUGUGCGCCGUUGCCGAUUUUCACUCACUGAGUGAGCCAAGUCCAUUGGAAUCAAGUUGCUCAACGGCAACGUUUCAUUU ...(((((....-----.....)))))..((((((((((((((((((...)))))))((((.....))))))))..(((....))))))))))........ ( -29.70) >DroYak_CAF1 31277 96 - 1 CCUGCACACACG-----GCUCUUGUGCGCCGUUGCCAAUUUUCACUCACUGAGUGAGCCGCGUCCAUUGGAAUCAAGUUGCUCAACGGCAACGUUUCAUUU ...(((((....-----.....)))))..(((((((.......((((...))))((((.((.(((...))).....)).))))...)))))))........ ( -27.90) >DroAna_CAF1 30482 99 - 1 CCUGCACACACGAUGGGGCU-AGGGGCAGCGUUGCCGAUUUUCAGUUACUGAGUGAGAAAAGCGGCCUGGAAGCCAGUUGCUCAACGGCAACGUUUCAUC- (((((.((.....))..)).-)))((.((((((((((...(((((...)))))((((...((((((......))).))).)))).))))))))))))...- ( -33.80) >consensus CCUGCACACACG_____GCUCUUGUGCGCCGUUGCCGAUUUUCACUCACUGAGUGAGCCAAGUCCAUUGGAAUCAAGUUGCUCAACGGCAACGUUUCAUUU ...((((................))))..(((((((....(((((((...)))))))...................(((....))))))))))........ (-19.14 = -19.64 + 0.50)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:08:52 2006