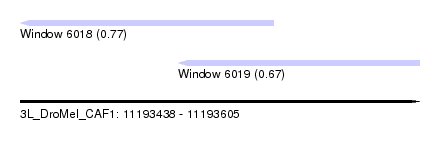
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 11,193,438 – 11,193,605 |
| Length | 167 |
| Max. P | 0.770185 |

| Location | 11,193,438 – 11,193,544 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 84.22 |
| Mean single sequence MFE | -29.63 |
| Consensus MFE | -20.66 |
| Energy contribution | -21.97 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.01 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.770185 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
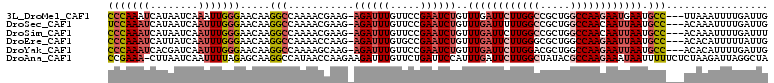
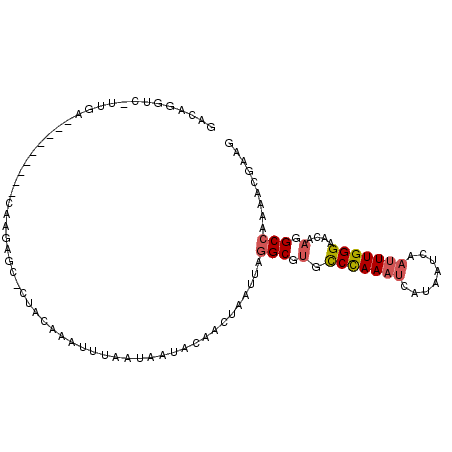
>3L_DroMel_CAF1 11193438 106 - 23771897 CCCAAAUCAUAAUCAAAUUGGGAACAAGGCCAAAACGAAG-AGAUUUGUUCCGAAUCUGUUUGAUUCUUGGCCGCUGGCCAAGAAUGAAUGCC---UUAAAUUUUGAUUG .........((((((((..(....)(((((.....((((.-(((((((...))))))).))))((((((((((...))))))))))....)))---))....)))))))) ( -34.70) >DroSec_CAF1 2585 106 - 1 UCCAAAUCAUAAUCAAUUUGGGAACAAGGCCAAAACGAAG-AGAUUUGUUCCGAAUCUGUUUGAUUUUUGGCCGCUGGCCAACAAUUAAUGCC---ACAAAUUUUGAUUG .........(((((((((((.......((((((((((((.-(((((((...))))))).))))..))))))))..((((...........)))---))))))..)))))) ( -29.70) >DroSim_CAF1 2426 106 - 1 CCCAAAUCAUAAUCAAUUUGGGAACAAGGCCAAAACGAAG-AGAUUUGUUCCGAAUCUGUUUGAUUCUUGGCCGCUGGCCAACAAUUAAUGCC---ACAAAUUUUGAUUU (((((((........)))))))..(((((((((((((((.-(((((((...))))))).)))).)).))))))(.((((...........)))---)).....))).... ( -30.00) >DroEre_CAF1 2846 106 - 1 CCCAAAUCAUUAUCAAUUUGGGAACAAGGCCAAAACCAAG-AGAUUUGUGCCGAAUCUGUUUGAUUCUUGGGCGCUGGCCAAGAAUUAAUGCC---ACACAUUUUUAUUG (((((((........))))))).....(((..........-(((((((...)))))))..(((((((((((.(...).))))))))))).)))---.............. ( -28.90) >DroYak_CAF1 2535 106 - 1 CCCAAAUCACGAUCAAUUUGGGAACAAGGCCAAAAGCAAG-AGAUUUGUUCCGAAUCUGUUUGAUUCUUGGACGCUGGCCAAGAAUUAAUGCC---ACACAUUUUGAUUG .........(((((((..((((((((((.((........)-.).))))))))........(((((((((((.(....))))))))))))....---...))..))))))) ( -28.70) >DroAna_CAF1 2391 109 - 1 CCGAAA-CUUAAUCAAUUUUAGAGCAAGGCCAUAACCAAGAAGAUUUGUUCUGAUUCCAUUUGAUUCUUGGCUAUACGCCAAGAAAUAAUUUUUCUCUAAGAUUAGGCUA ......-(((((((....((((((((((................))))))))))..........((((((((.....))))))))...............)))))))... ( -25.79) >consensus CCCAAAUCAUAAUCAAUUUGGGAACAAGGCCAAAACCAAG_AGAUUUGUUCCGAAUCUGUUUGAUUCUUGGCCGCUGGCCAAGAAUUAAUGCC___ACAAAUUUUGAUUG (((((((........))))))).....(((...........((((((.....))))))..((((((((((((.....)))))))))))).)))................. (-20.66 = -21.97 + 1.31)
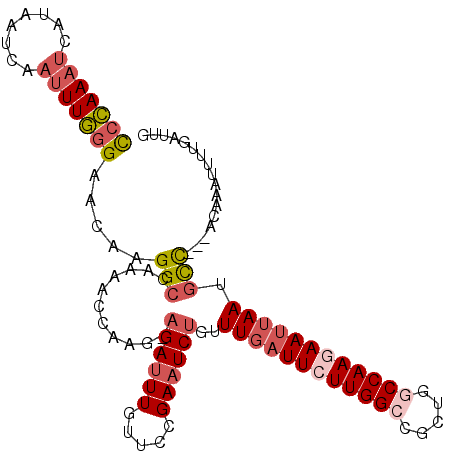
| Location | 11,193,504 – 11,193,605 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 78.54 |
| Mean single sequence MFE | -20.28 |
| Consensus MFE | -10.48 |
| Energy contribution | -10.87 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.670485 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11193504 101 - 23771897 GACAGGUC-UUGACGGCAAAAUCAAGAAC-CUACAAAUUUAAUAAUACAACUAAUUGGGCGUGCCCAAAUCAUAAUCAAAUUGGGAACAAGGCCAAAACGAAG ...(((((-((((........))))).))-))......................((((.(...(((((............))))).....).))))....... ( -19.80) >DroPse_CAF1 2480 90 - 1 GACAUUCCCCUGA---------CAAGAAACACCCAAAUUU--UGAU-CAACCAAUUAAGCAGGCCUAAA-CACGAUCAAUUUGGGAACAAGGCCACAGCGGAG .....(((.(((.---------(((((..........)))--))..-..............(((((...-..(((.....)))......))))).))).))). ( -16.70) >DroSec_CAF1 2651 86 - 1 ----GGUC-UA-----------CAAGAGC-CUACAAAUUGAAUAAUACAACUAAUUGGGCGUGUCCAAAUCAUAAUCAAUUUGGGAACAAGGCCAAAACGAAG ----((((-(.-----------.....((-(((....(((.......))).....)))))((..((((((........))))))..)).)))))......... ( -19.60) >DroSim_CAF1 2492 90 - 1 GACAGGUC-UU-----------CAAGAGC-CUACAAAUUGAAUAAUACAACUAAUUGGGCGUGCCCAAAUCAUAAUCAAUUUGGGAACAAGGCCAAAACGAAG ....((((-((-----------.....((-(((....(((.......))).....)))))((.(((((((........))))))).))))))))......... ( -22.40) >DroEre_CAF1 2912 101 - 1 GACAGGUC-UUGACGGCAUAUUCAAGAGC-CCACAAAUUUAAUAAUACAACUAAUUAGGCAUGCCCAAAUCAUUAUCAAUUUGGGAACAAGGCCAAAACCAAG ....((((-(((..(((..........))-)................................(((((((........)))))))..)))))))......... ( -21.60) >DroYak_CAF1 2601 101 - 1 GACAGGUC-GUGACGGCAUAUUCAAGAGC-CAACAAAUUUAAUAAUACAACUAAUUAGGCGUGCCCAAAUCACGAUCAAUUUGGGAACAAGGCCAAAAGCAAG ....((((-((((.(((((........((-(..........................))))))))....))))))))..(((((........)))))...... ( -21.57) >consensus GACAGGUC_UUGA_________CAAGAGC_CUACAAAUUUAAUAAUACAACUAAUUAGGCGUGCCCAAAUCAUAAUCAAUUUGGGAACAAGGCCAAAACGAAG .........................................................(((.(.(((((((........)))))))....).)))......... (-10.48 = -10.87 + 0.39)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:08:42 2006