
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 11,179,218 – 11,179,317 |
| Length | 99 |
| Max. P | 0.803009 |

| Location | 11,179,218 – 11,179,317 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 91.19 |
| Mean single sequence MFE | -25.30 |
| Consensus MFE | -19.78 |
| Energy contribution | -20.22 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.803009 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
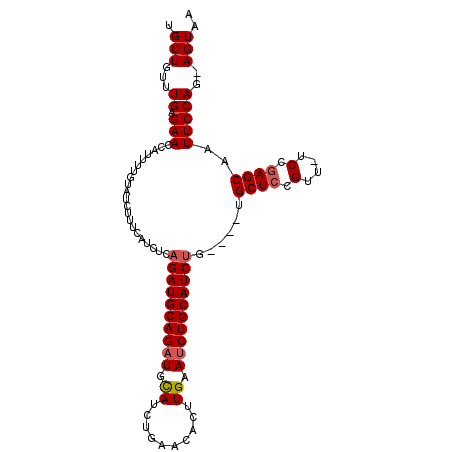
>3L_DroMel_CAF1 11179218 99 + 23771897 UGCUGUUUGAGAACCAUUUUGUAUCUUUCAUCUCAGAUGCAGAUGCAUCUGAACACUUGAAUCUGCAUCU------GCUCCGUUUUCCGAGCAAUUCCAG-AGUAA ((((..(((.(((......((((...((((..((((((((....)))))))).....))))..))))..(------((((.(....).))))).))))))-)))). ( -24.90) >DroSec_CAF1 39211 100 + 1 UGCUGUUUGAGAACCAUUUUGUAUCUUUCAUCUCAGAUGCAGAUGCAUCUGAACACUUGAAUCUGCAUCUG----UGCUCCGUU-UCCGAGCAAUUCCAG-AGUAA (((.(((..((.((......))..........((((((((....))))))))...))..)))..)))((((----(((((....-...)))))....)))-).... ( -26.20) >DroSim_CAF1 39029 101 + 1 UGCUGUUUGAGAACCAUUUUGUAUCUUUCAUCUCUGAUGCAGAUGCAUCUGAACACUUGAAGCUGCAUCUG----UGCUCCGUG-UCCGAGCAAUUCCAGAAGUAA ((((...(((((.............)))))..((((..(((((((((.((..........)).))))))))----)((((....-...)))).....)))))))). ( -24.72) >DroEre_CAF1 40718 98 + 1 GGCUAUUUGAGAACCAUUUUGUAUCUUGCAUCUCAGAUGCAGAUGCAUCUGAGUACUUGAAUCUGCAUCU------GCUCCGUU-UCCGAGCAAUUCCAG-AGUAA .....((..((.((((...(((((((.(((((...))))))))))))..)).)).))..))((((.(..(------((((....-...)))))..).)))-).... ( -28.40) >DroYak_CAF1 41968 104 + 1 GGCUAUUUGAGAACCACUUUGUAUCUUUCAUCUGAGAUGCAGAUGUAUCUGAAUACUUGAAUCUGCAUCUGCAACUGCUCCGUU-UCCCAGCAAUUCCAG-AGUAA ...(((((.(((..((.((((((((((......))))))))))))..))))))))........(((.((((.((.((((.....-....)))).)).)))-)))). ( -22.30) >consensus UGCUGUUUGAGAACCAUUUUGUAUCUUUCAUCUCAGAUGCAGAUGCAUCUGAACACUUGAAUCUGCAUCUG____UGCUCCGUU_UCCGAGCAAUUCCAG_AGUAA .(((...((.(((.....................((((((((((.((..........)).))))))))))......((((.(....).))))..)))))..))).. (-19.78 = -20.22 + 0.44)


| Location | 11,179,218 – 11,179,317 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 91.19 |
| Mean single sequence MFE | -28.27 |
| Consensus MFE | -19.66 |
| Energy contribution | -20.06 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.580859 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11179218 99 - 23771897 UUACU-CUGGAAUUGCUCGGAAAACGGAGC------AGAUGCAGAUUCAAGUGUUCAGAUGCAUCUGCAUCUGAGAUGAAAGAUACAAAAUGGUUCUCAAACAGCA ....(-(((.(.((((((.(....).))))------)).).))))((((....(((((((((....))))))))).)))).......................... ( -28.40) >DroSec_CAF1 39211 100 - 1 UUACU-CUGGAAUUGCUCGGA-AACGGAGCA----CAGAUGCAGAUUCAAGUGUUCAGAUGCAUCUGCAUCUGAGAUGAAAGAUACAAAAUGGUUCUCAAACAGCA .....-(((....((((((..-..).)))))----(((((((((((.((..........)).))))))))))).((.(((..((.....))..)))))...))).. ( -28.40) >DroSim_CAF1 39029 101 - 1 UUACUUCUGGAAUUGCUCGGA-CACGGAGCA----CAGAUGCAGCUUCAAGUGUUCAGAUGCAUCUGCAUCAGAGAUGAAAGAUACAAAAUGGUUCUCAAACAGCA .............((((.(((-((((((((.----........)))))..)))))).(((((....))))).((((..................))))....)))) ( -23.97) >DroEre_CAF1 40718 98 - 1 UUACU-CUGGAAUUGCUCGGA-AACGGAGC------AGAUGCAGAUUCAAGUACUCAGAUGCAUCUGCAUCUGAGAUGCAAGAUACAAAAUGGUUCUCAAAUAGCC ....(-(((.(.(((((((..-..).))))------)).).)))).....((((((((((((....))))))))).)))............((((.......)))) ( -33.30) >DroYak_CAF1 41968 104 - 1 UUACU-CUGGAAUUGCUGGGA-AACGGAGCAGUUGCAGAUGCAGAUUCAAGUAUUCAGAUACAUCUGCAUCUCAGAUGAAAGAUACAAAGUGGUUCUCAAAUAGCC .....-.((((((..((.(..-..)....((.(((.((((((((((....((((....))))))))))))))))).))..........))..)))).))....... ( -27.30) >consensus UUACU_CUGGAAUUGCUCGGA_AACGGAGCA____CAGAUGCAGAUUCAAGUGUUCAGAUGCAUCUGCAUCUGAGAUGAAAGAUACAAAAUGGUUCUCAAACAGCA .......(((((((((((.(....).))))......((((((((((....((((....))))))))))))))...................))))).))....... (-19.66 = -20.06 + 0.40)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:08:36 2006