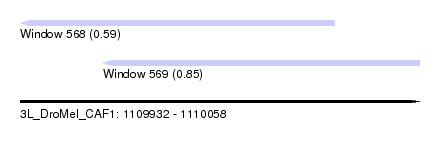
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,109,932 – 1,110,058 |
| Length | 126 |
| Max. P | 0.846158 |

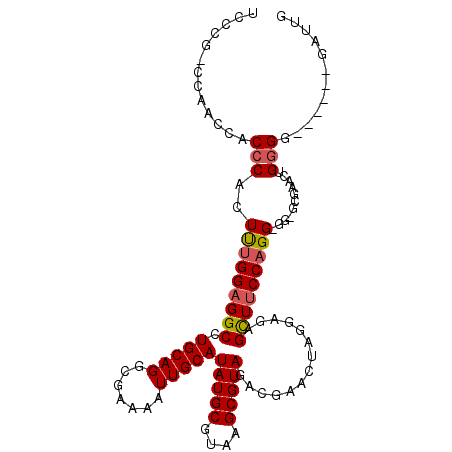
| Location | 1,109,932 – 1,110,031 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.20 |
| Mean single sequence MFE | -32.62 |
| Consensus MFE | -16.55 |
| Energy contribution | -17.61 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.589391 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1109932 99 - 23771897 UGCAGGCGAAAAUUGCAUAUGCGUAAGCGUAGACGAACUAGGAGAGCUUCCAGGGGG-GCGAACUGGGG------GAUUGGGGUGGCU--UUCCGAGAGGGUUCUAGG------------ (((((.......)))))(((((....)))))......((((((...((((..((..(-((.(.(..(..------..)..)..).)))--..))..)))).)))))).------------ ( -32.50) >DroSec_CAF1 3456 98 - 1 UGCAGGCGAAAAUUGCAUAUGCGUAAGCGUAGACGAACUAGGAGAGCUUCCAGG-GG-GCGAACUGGGG------GAUUGGGGUGGCU--UUCCAAGAGGGUUCUAGG------------ (((((.......)))))(((((....)))))...(((((.(((((((((((((.-..-.....))))).------.........))))--)))).....)))))....------------ ( -30.60) >DroSim_CAF1 995 98 - 1 UGCAGGCGAAAAUUGCAUAUGCGUAAGCGUAGACGAACUAGGAGAGCUUCCAGG-GG-GCGAACUGGGG------GAUUGGGGUGGCU--UUCCGAGAGGGUUCUAGG------------ (((((.......)))))(((((....)))))...(((((.(((((((((((((.-..-.....))))).------.........))))--)))).....)))))....------------ ( -31.70) >DroEre_CAF1 1481 97 - 1 UGCAGGCGAAAAUUGCAUAUGCGUAAGCGUAGACGA------GGAGUUUCCAGG-GG--GCGAUGGGGAAGCUGGGAGAGGGGUGGCU--UUCCGAGAGGGUUCUAGG------------ (((((.......)))))(((((....)))))..(.(------(((.(..(((..-..--....)))..)..((.((((((......))--)))).))....)))).).------------ ( -26.50) >DroYak_CAF1 3389 114 - 1 UGCAGGCGAAAAUUGCAUAUGCGUAAGCGUAGACGAACUAGGAGAGCUUCCAGG-GG--CAAACCGGGGAACUG-GAUUGGGGUGGCU--UUCCGAGAGGGUUCUAGGGGCUGGGGGCUA .((.(((.......(..(((((....)))))..)(((((.((((((((.((...-..--(((.((((....)))-).))).)).))))--)))).....))))).....)))....)).. ( -40.70) >DroAna_CAF1 3751 95 - 1 UGCAGGCGAAAAUUGCAUAUGCGUAAGCGUAGACGAACCAGGAAGGUUACCAGG-AAGGUGG---GUGGUGCUG-GGCAGGAGCGGCCUGUUUCGAAAGG-------------------- .((((((.....((((.(((((....)))))......((((.(.(.(((((...-..)))))---.)..).)))-))))).....))))))..(....).-------------------- ( -33.70) >consensus UGCAGGCGAAAAUUGCAUAUGCGUAAGCGUAGACGAACUAGGAGAGCUUCCAGG_GG_GCGAACUGGGG______GAUUGGGGUGGCU__UUCCGAGAGGGUUCUAGG____________ (((((.......)))))(((((....)))))...(((((.(((((((((((((........................)))))..))))..)))).....)))))................ (-16.55 = -17.61 + 1.06)

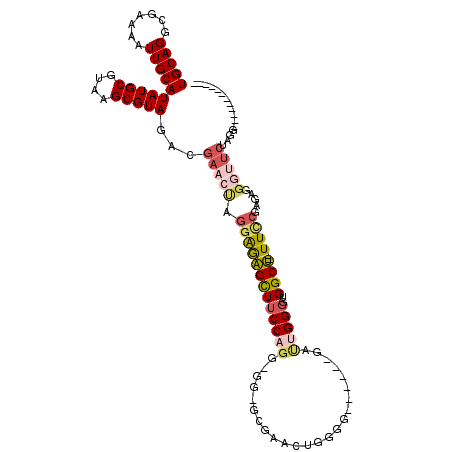
| Location | 1,109,958 – 1,110,058 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 78.10 |
| Mean single sequence MFE | -38.30 |
| Consensus MFE | -21.20 |
| Energy contribution | -21.34 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.846158 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1109958 100 - 23771897 UCCCG-CCAACCACCCACUUUGGAGGCCUGCAGGCGAAAAUUGCAUAUGCGUAAGCGUAGACGAACUAGGAGAGCUUCCAGGGGG-GCGAACUGGGG------GAUUG ((((.-(((..(.(((.((((((((((((.(((((((...)))).(((((....)))))......)).).)).))))))))))))-).)...)))))------))... ( -43.10) >DroSec_CAF1 3482 100 - 1 UCCCGCCCAGCCACCCACUUUGGAGGCCUGCAGGCGAAAAUUGCAUAUGCGUAAGCGUAGACGAACUAGGAGAGCUUCCAGG-GG-GCGAACUGGGG------GAUUG ..((.(((((....((.((((((((((((.(((((((...)))).(((((....)))))......)).).)).)))))))))-))-)....))))))------).... ( -41.10) >DroSim_CAF1 1021 100 - 1 UCCCGACCAGCCACCCACUUUGGAGGCCUGCAGGCGAAAAUUGCAUAUGCGUAAGCGUAGACGAACUAGGAGAGCUUCCAGG-GG-GCGAACUGGGG------GAUUG ((((..((((....((.((((((((((((.(((((((...)))).(((((....)))))......)).).)).)))))))))-))-)....))))))------))... ( -39.90) >DroEre_CAF1 1507 90 - 1 ACC---------ACCCACUUUGGAGGCCUGCAGGCGAAAAUUGCAUAUGCGUAAGCGUAGACGA------GGAGUUUCCAGG-GG--GCGAUGGGGAAGCUGGGAGAG ...---------.(((.((((((((((((((((.......)))).(((((....))))).....------)).)))))))))-)(--((.........)))))).... ( -30.60) >DroYak_CAF1 3427 98 - 1 ACC------ACCACCCACUUUGGAGGCCUGCAGGCGAAAAUUGCAUAUGCGUAAGCGUAGACGAACUAGGAGAGCUUCCAGG-GG--CAAACCGGGGAACUG-GAUUG ...------........((((((((((((.(((((((...)))).(((((....)))))......)).).)).)))))))))-).--(((.((((....)))-).))) ( -34.60) >DroAna_CAF1 3771 103 - 1 UCCUGUGAGCCCUCCCCCUCCGGAGGCCUGCAGGCGAAAAUUGCAUAUGCGUAAGCGUAGACGAACCAGGAAGGUUACCAGG-AAGGUGG---GUGGUGCUG-GGCAG ..((((.(((((.((((((((((..((((((((.......)))).(((((....)))))............))))..)).))-.))).))---).)).))).-).))) ( -40.50) >consensus UCCCG_CCAACCACCCACUUUGGAGGCCUGCAGGCGAAAAUUGCAUAUGCGUAAGCGUAGACGAACUAGGAGAGCUUCCAGG_GG_GCGAACUGGGG______GAUUG .............(((..(((((((((.(((((.......)))))(((((....)))))..............)))))))))...........)))............ (-21.20 = -21.34 + 0.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:40:40 2006