
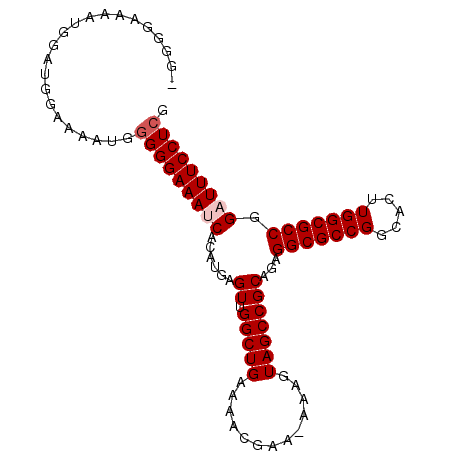
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 11,165,698 – 11,165,795 |
| Length | 97 |
| Max. P | 0.999942 |

| Location | 11,165,698 – 11,165,795 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 94.64 |
| Mean single sequence MFE | -31.50 |
| Consensus MFE | -29.48 |
| Energy contribution | -30.08 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.94 |
| SVM decision value | 4.41 |
| SVM RNA-class probability | 0.999891 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11165698 97 + 23771897 GGGGAAAAAUGGAUGGAAAAUGGGGGAAAUCACAUGAGUUGGCUGAAAACGAA-AAAGUAGCCGCAGAGGCGCCGGCACUUGGCGCCGGAUUUCCUCG ......................(((((((((......((.(((((........-....)))))))...(((((((.....))))))).))))))))). ( -32.30) >DroSec_CAF1 26106 97 + 1 GGGGGAAAAUGGAUGGAAAAUGGGGGAAAACACAUGAGUUGGCUGAAAACGAA-AAAGUAGCCGCAGAGGCGCCGGCACUUGGCGCCGGAUUUCCUCG .....................(((((((..(......((.(((((........-....)))))))...(((((((.....))))))))..))))))). ( -28.50) >DroSim_CAF1 26850 95 + 1 -GGGGAAAAUGGAUGGA-AAUGGGGGAAAACACAUGAGUUGGCUGAAAACGAA-AAAGUAGCCGCAGAGGCGCCGGCACUUGGCGCCGGAUUUCCUCG -................-...(((((((..(......((.(((((........-....)))))))...(((((((.....))))))))..))))))). ( -28.50) >DroEre_CAF1 27469 97 + 1 -GGGGAAAGCGGAUGGAAAAUGUGGGAAAUCACAUGAGUUGGCUGAAAAGGAAAAAAGUAGCCGCAGAGGCGCCGGCACUUGGCGCCGGAUUUCCUCA -(((((((((((.......((((((....)))))).......((....))...........))))...(((((((.....)))))))...))))))). ( -35.70) >DroYak_CAF1 27844 96 + 1 -GGGGAAAAUGGUUGGAAAAUGGGGGAAAUCACAUGAGUUGGCUGAAAAGGAA-AAAGUAGCCGCAGAGGCGCCGGCACUUGGCGCCGGAUUUCCUCA -..........(((....))).(((((((((......((.(((((........-....)))))))...(((((((.....))))))).))))))))). ( -32.50) >consensus _GGGGAAAAUGGAUGGAAAAUGGGGGAAAUCACAUGAGUUGGCUGAAAACGAA_AAAGUAGCCGCAGAGGCGCCGGCACUUGGCGCCGGAUUUCCUCG ......................(((((((((......((.(((((.............)))))))...(((((((.....))))))).))))))))). (-29.48 = -30.08 + 0.60)


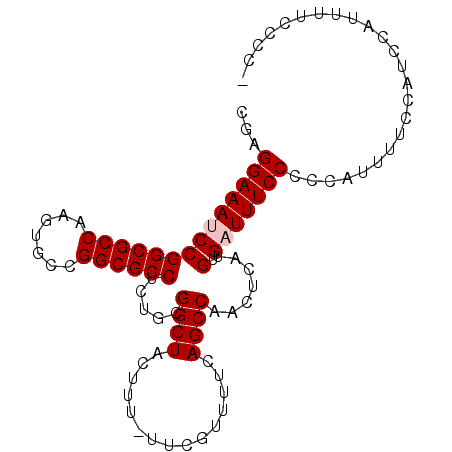
| Location | 11,165,698 – 11,165,795 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 94.64 |
| Mean single sequence MFE | -24.90 |
| Consensus MFE | -23.86 |
| Energy contribution | -24.26 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.96 |
| SVM decision value | 4.71 |
| SVM RNA-class probability | 0.999942 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11165698 97 - 23771897 CGAGGAAAUCCGGCGCCAAGUGCCGGCGCCUCUGCGGCUACUUU-UUCGUUUUCAGCCAACUCAUGUGAUUUCCCCCAUUUUCCAUCCAUUUUUCCCC ...((((((((((((((.......)))))).....((((((...-...))....)))).......).)))))))........................ ( -25.90) >DroSec_CAF1 26106 97 - 1 CGAGGAAAUCCGGCGCCAAGUGCCGGCGCCUCUGCGGCUACUUU-UUCGUUUUCAGCCAACUCAUGUGUUUUCCCCCAUUUUCCAUCCAUUUUCCCCC .((((....(((((((...)))))))..))))...((((((...-...))....))))........................................ ( -22.40) >DroSim_CAF1 26850 95 - 1 CGAGGAAAUCCGGCGCCAAGUGCCGGCGCCUCUGCGGCUACUUU-UUCGUUUUCAGCCAACUCAUGUGUUUUCCCCCAUU-UCCAUCCAUUUUCCCC- ...((((((..((((((.......)))))).....((((((...-...))....))))...................)))-))).............- ( -23.10) >DroEre_CAF1 27469 97 - 1 UGAGGAAAUCCGGCGCCAAGUGCCGGCGCCUCUGCGGCUACUUUUUUCCUUUUCAGCCAACUCAUGUGAUUUCCCACAUUUUCCAUCCGCUUUCCCC- .((((....(((((((...)))))))..)))).((((((...............)))).....(((((......))))).........)).......- ( -27.36) >DroYak_CAF1 27844 96 - 1 UGAGGAAAUCCGGCGCCAAGUGCCGGCGCCUCUGCGGCUACUUU-UUCCUUUUCAGCCAACUCAUGUGAUUUCCCCCAUUUUCCAACCAUUUUCCCC- ...((((((((((((((.......)))))).....((((.....-.........)))).......).))))))).......................- ( -25.74) >consensus CGAGGAAAUCCGGCGCCAAGUGCCGGCGCCUCUGCGGCUACUUU_UUCGUUUUCAGCCAACUCAUGUGAUUUCCCCCAUUUUCCAUCCAUUUUCCCC_ ...((((((((((((((.......)))))).....((((...............)))).......).)))))))........................ (-23.86 = -24.26 + 0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:08:27 2006