| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 11,164,333 – 11,164,432 |
| Length | 99 |
| Max. P | 0.732816 |

| Location | 11,164,333 – 11,164,432 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 76.21 |
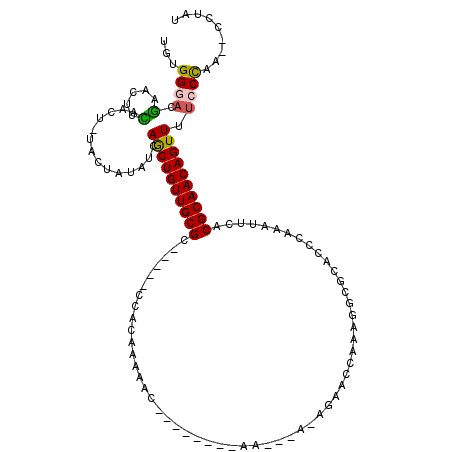
| Mean single sequence MFE | -21.08 |
| Consensus MFE | -11.60 |
| Energy contribution | -11.36 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.732816 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
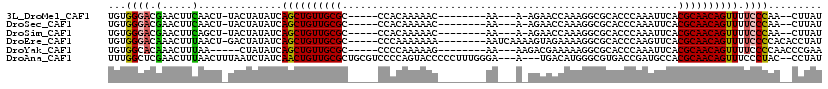
>3L_DroMel_CAF1 11164333 99 - 23771897 UGUGGGACGAACUUCAACU-UACUAUAUCAGCUGUUGCGC-----CCACAAAAAC--------AA---A-AGAACCAAAGGCGCACCCAAAUUCACGCAACAGUUUUCCCAA--CUUAU ..(((((.(.....)....-.........(((((((((((-----(.........--------..---.-.........)))(....)........))))))))).))))).--..... ( -18.40) >DroSec_CAF1 24797 99 - 1 UGUGGGACGAACUUCAACU-UACUAUAUCAGCUGUUGCGC-----CCACAAAAAC--------AA---A-AGAACCAAAGGCGCACCCAAAUUCACGCAACAGUUUUCCCAA--CUUAU ..(((((.(.....)....-.........(((((((((((-----(.........--------..---.-.........)))(....)........))))))))).))))).--..... ( -18.40) >DroSim_CAF1 25496 99 - 1 UGUGGGACGAACUUCAGCU-UACUAUAUCAGCUGUUGCGC-----CCACAAAAAC--------AA---A-AGAACCAAAGGCGCACCCAAAUUCACGCAACAGUUUUCCCAA--CUUAU ..(((((.(((((.(((((-.........))))).(((((-----(.........--------..---.-.........))))))................)))))))))).--..... ( -22.80) >DroEre_CAF1 26013 105 - 1 UGUGGGACAAACUUUAACU-GACUAUAUCAGCUGUUGCGC-----CCCAAAAAAA--------AAUCAAAAGUAGAAAAGGCGCACCCAAGUUCACGCAACAGUUUUCCCCACACCUAU ((((((...........((-((.....))))(((((((((-----(.........--------................)))((......))....))))))).....))))))..... ( -23.51) >DroYak_CAF1 26300 98 - 1 UGUGGCACAAACUUUAA-----CUAUAUCAGCUGUUGCGC-----CCCCAAAAAG--------AA---AAGACGAAAAAGGCGCACCCAAAUUCACGCAACAGUUUUCCCCAACCCGAA .................-----.......((((((((((.-----.........(--------..---....)(((...((.....))...))).)))))))))).............. ( -13.10) >DroAna_CAF1 27616 111 - 1 UUUGGCUCGAACUUUAACUUUAAUCUAUCAACUGUUGCGCUGCGUCCCCAGUACCCCCUUUGGGA---A---UGACAUGGGCGUGACCGAUGCCACGCAACAGUUUCCCUAC--CCUAU .............................((((((((((..((((((((((........))))).---.---......((......)))))))..)))))))))).......--..... ( -30.30) >consensus UGUGGGACGAACUUCAACU_UACUAUAUCAGCUGUUGCGC_____CCACAAAAAC________AA___A_AGAACCAAAGGCGCACCCAAAUUCACGCAACAGUUUUCCCAA__CCUAU ...((((.(.....)..............((((((((((........................................................)))))))))).))))......... (-11.60 = -11.36 + -0.25)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:08:25 2006