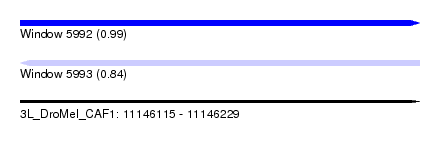
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 11,146,115 – 11,146,229 |
| Length | 114 |
| Max. P | 0.988165 |

| Location | 11,146,115 – 11,146,229 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.29 |
| Mean single sequence MFE | -27.45 |
| Consensus MFE | -16.34 |
| Energy contribution | -16.98 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.60 |
| SVM decision value | 2.11 |
| SVM RNA-class probability | 0.988165 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11146115 114 + 23771897 ACUAACUUAUUCUCAGAAAUAUGUAACCUUGUAUGUGUUCAACUUGAUGGGCGAAAAUCCCGUCGAACCAUGACAAUAUGAAUGUAAUUAUUUUAUAAUAUGACAGAGAAUGAG------ .....(((((((((..............(((((((.((((.....((((((.......))))))))))))).))))......(((...((((....))))..))))))))))))------ ( -26.10) >DroSec_CAF1 7065 120 + 1 ACUAACUUAUUCUCAGAAAUAUGUAACCUUGUAAGUGUUCAACUUGAUAGGCGAAAAUCCAGUCGAACCAUGACAAUAUGAAUGUAAUUGUUUUAUAAUAUGACAGAGAAUAAGUUGGAG .(((((((((((((..............((((....((((.(((.(((........))).))).))))....))))......(((...((((....))))..)))))))))))))))).. ( -29.30) >DroSim_CAF1 7113 119 + 1 ACUAACUUAUUCUCAGAAAUCGGUAACCUUGUA-GUGUUCAACUUGAUAGGCGAAAUUCCCGUCGAACCAUGACAAUAUGAAUGUAAUUGUUUUAUAAUAUGACAGAGAAUAAGUUGGAG .(((((((((((((.....(((....(((...(-((.....)))....)))))).......((((......((((((.........))))))........)))).))))))))))))).. ( -28.04) >DroEre_CAF1 7132 104 + 1 ACUAACUUGUUCUCAGAAAUAUUUAACCUUG----UGUUCAACU-------CGAAAAGCCCUUCGAACCAUGACAAUAUGAAUGCAGUUGUUAUAUAAU-----AGAGAAUGAGUUGGGG .((((((..(((((.....(((.((((.(((----((((((..(-------((((......)))))...((....)).)))))))))..)))).)))..-----.)))))..)))))).. ( -27.40) >DroYak_CAF1 7408 104 + 1 ACCAACUUAUUCUUAGAAAUAUUUAACCUUG----UGUUCAAAU-------CGAAAACACCUUCCAUCCAUGACAAUAUGAAUGCAGUUGUUAAAUUAU-----ACAGAAUAAGUUGGGU .((((((((((((.......(((((((.(((----((((((...-------.(((......)))((....))......)))))))))..)))))))...-----..)))))))))))).. ( -26.40) >consensus ACUAACUUAUUCUCAGAAAUAUGUAACCUUGUA_GUGUUCAACUUGAU_GGCGAAAAUCCCGUCGAACCAUGACAAUAUGAAUGUAAUUGUUUUAUAAUAUGACAGAGAAUAAGUUGGAG .(((((((((((((.....................................(((........)))......((((((.........)))))).............))))))))))))).. (-16.34 = -16.98 + 0.64)


| Location | 11,146,115 – 11,146,229 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.29 |
| Mean single sequence MFE | -24.68 |
| Consensus MFE | -13.19 |
| Energy contribution | -14.35 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.843738 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11146115 114 - 23771897 ------CUCAUUCUCUGUCAUAUUAUAAAAUAAUUACAUUCAUAUUGUCAUGGUUCGACGGGAUUUUCGCCCAUCAAGUUGAACACAUACAAGGUUACAUAUUUCUGAGAAUAAGUUAGU ------((.((((((.(..((((.....................((((.((((((((((..(((........)))..))))))).)))))))(....)))))..).)))))).))..... ( -21.00) >DroSec_CAF1 7065 120 - 1 CUCCAACUUAUUCUCUGUCAUAUUAUAAAACAAUUACAUUCAUAUUGUCAUGGUUCGACUGGAUUUUCGCCUAUCAAGUUGAACACUUACAAGGUUACAUAUUUCUGAGAAUAAGUUAGU ....(((((((((((.(..((((.....................((((....((((((((.(((........))).))))))))....))))(....)))))..).)))))))))))... ( -26.80) >DroSim_CAF1 7113 119 - 1 CUCCAACUUAUUCUCUGUCAUAUUAUAAAACAAUUACAUUCAUAUUGUCAUGGUUCGACGGGAAUUUCGCCUAUCAAGUUGAACAC-UACAAGGUUACCGAUUUCUGAGAAUAAGUUAGU ....(((((((((((.(((.........................((((....(((((((..((..........))..)))))))..-.))))((...)))))....)))))))))))... ( -25.70) >DroEre_CAF1 7132 104 - 1 CCCCAACUCAUUCUCU-----AUUAUAUAACAACUGCAUUCAUAUUGUCAUGGUUCGAAGGGCUUUUCG-------AGUUGAACA----CAAGGUUAAAUAUUUCUGAGAACAAGUUAGU ....((((..(((((.-----..(((.((((...((..((((..(((....((((.....))))...))-------)..))))..----))..)))).))).....)))))..))))... ( -17.60) >DroYak_CAF1 7408 104 - 1 ACCCAACUUAUUCUGU-----AUAAUUUAACAACUGCAUUCAUAUUGUCAUGGAUGGAAGGUGUUUUCG-------AUUUGAACA----CAAGGUUAAAUAUUUCUAAGAAUAAGUUGGU ..((((((((((((..-----...(((((((..((.(((((((......)))))))..))((((((...-------....)))))----)...))))))).......)))))))))))). ( -32.30) >consensus CCCCAACUUAUUCUCUGUCAUAUUAUAAAACAAUUACAUUCAUAUUGUCAUGGUUCGACGGGAUUUUCGCC_AUCAAGUUGAACAC_UACAAGGUUACAUAUUUCUGAGAAUAAGUUAGU ....(((((((((((...............((...(((.......)))..))(((((((..................)))))))......................)))))))))))... (-13.19 = -14.35 + 1.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:08:17 2006