| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 11,145,588 – 11,145,691 |
| Length | 103 |
| Max. P | 0.572770 |

| Location | 11,145,588 – 11,145,691 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 81.50 |
| Mean single sequence MFE | -28.15 |
| Consensus MFE | -19.74 |
| Energy contribution | -19.38 |
| Covariance contribution | -0.35 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.70 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.524288 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
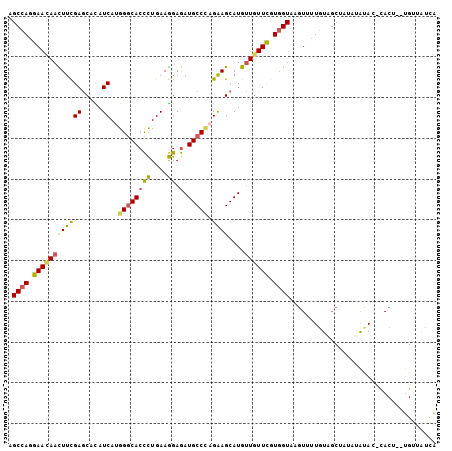
>3L_DroMel_CAF1 11145588 103 + 23771897 AGCCAGGAAUAAUUUCGAGCACAUCAUGGGCACCCUGAAGGAGAUGCCCAGAAGCAUGUUGUUCGUGGUAAGUUUUGUAGCUAUAUGCAUAUACU--UGUUAUCA .((((.(((((((.....((...((.(((((((((....)).).)))))))).))..))))))).))))((((..((((......))))...)))--)....... ( -30.50) >DroSec_CAF1 6541 103 + 1 AGCCAGGAACAACUUCGAGCACAUCAUGGGCACCCUGAAGGAGAUGCCCCGAAGCAUGUUGUUCGUGGUAAAUUUUGUAGCUAUAUAUACACACU--UGUUAUCA .((((.((((((((((((.....))..((((((((....)).).))))).)))....))))))).))))......((((........))))....--........ ( -28.10) >DroSim_CAF1 6589 103 + 1 AGCCAGGAACAACUUCGAGCACAUCAUGGGCACCCUGAAGGAGAUGCCCCGAAGCAUGUUGUUCGUGGUAAGUUUUGUAGCUAUAUAUACUCACU--UGUUAUCA .((((.((((((((((((.....))..((((((((....)).).))))).)))....))))))).))))((((...(((........)))..)))--)....... ( -29.10) >DroEre_CAF1 6612 101 + 1 AGCCAGGAACAACUUCGAGCAUAUCAUGGGCACCCUGAAGGAGAUGCCCAGGAGCAUGCUGUUCGUGGUGAGUGUUGUAGCAUUCCAUA--CACG--UGGUAUUA .((((.(((((.......((((.((.(((((((((....)).).)))))).))..))))))))).))))((((((....)))))).(((--(...--..)))).. ( -34.31) >DroYak_CAF1 6888 101 + 1 AGCCAGGAACAACUUCGAGCACAUCAUGGGCACCCUGAAGGAGAUGCCUAGGAGCAUGUUGUUCGUGGUAAGUGUUGUGGCAUUCUAUA--UACG--UGGUAUUA .((((.(((((((.....((.(....(((((((((....)).).)))))).).))..))))))).))))..((((.((((....)))))--))).--........ ( -30.60) >DroAna_CAF1 6239 101 + 1 AGCUAGGAAAAAUUUUGAACUUAUCAUGGACACUUUGAAAGAAAUGCCGAGAAGCAUGUUAUUUGUCGUAAGUUAUUUUUCUA---AUAC-CACUUUGGUUUUCC .....((((((((....(((((((....((((...(((.....((((......)))).)))..))))))))))))))))))).---..((-(.....)))..... ( -16.30) >consensus AGCCAGGAACAACUUCGAGCACAUCAUGGGCACCCUGAAGGAGAUGCCCAGAAGCAUGUUGUUCGUGGUAAGUUUUGUAGCUAUAUAUAC_CACU__UGUUAUCA .((((.((((((((((((.....))..((((((((....)).).))))).))))....)))))).)))).................................... (-19.74 = -19.38 + -0.35)

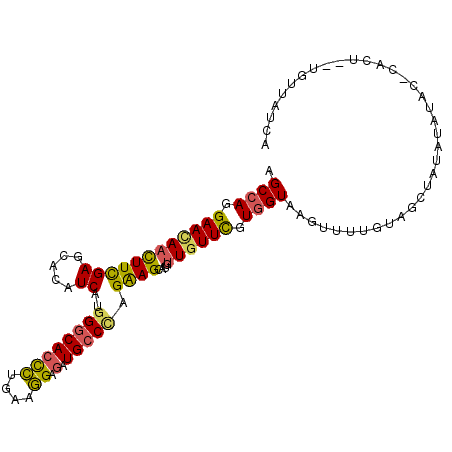
| Location | 11,145,588 – 11,145,691 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 81.50 |
| Mean single sequence MFE | -25.10 |
| Consensus MFE | -18.72 |
| Energy contribution | -19.33 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.572770 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
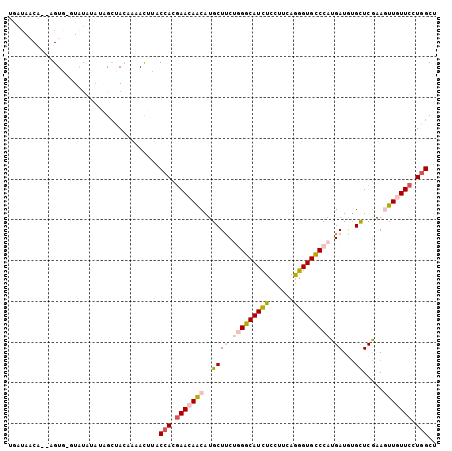
>3L_DroMel_CAF1 11145588 103 - 23771897 UGAUAACA--AGUAUAUGCAUAUAGCUACAAAACUUACCACGAACAACAUGCUUCUGGGCAUCUCCUUCAGGGUGCCCAUGAUGUGCUCGAAAUUAUUCCUGGCU .......(--(((....((.....))......)))).(((.(((......((.(((((((((((......))))))))).))...)).........))).))).. ( -21.66) >DroSec_CAF1 6541 103 - 1 UGAUAACA--AGUGUGUAUAUAUAGCUACAAAAUUUACCACGAACAACAUGCUUCGGGGCAUCUCCUUCAGGGUGCCCAUGAUGUGCUCGAAGUUGUUCCUGGCU ........--....((((........)))).......(((.(((((((..((.(((((((((((......)))))))).)))...)).....))))))).))).. ( -29.10) >DroSim_CAF1 6589 103 - 1 UGAUAACA--AGUGAGUAUAUAUAGCUACAAAACUUACCACGAACAACAUGCUUCGGGGCAUCUCCUUCAGGGUGCCCAUGAUGUGCUCGAAGUUGUUCCUGGCU .......(--(((..(((........)))...)))).(((.(((((((..((.(((((((((((......)))))))).)))...)).....))))))).))).. ( -30.30) >DroEre_CAF1 6612 101 - 1 UAAUACCA--CGUG--UAUGGAAUGCUACAACACUCACCACGAACAGCAUGCUCCUGGGCAUCUCCUUCAGGGUGCCCAUGAUAUGCUCGAAGUUGUUCCUGGCU ........--.(((--(.((........))))))...(((.((((((((((.((.(((((((((......))))))))).))))))).......))))).))).. ( -32.21) >DroYak_CAF1 6888 101 - 1 UAAUACCA--CGUA--UAUAGAAUGCCACAACACUUACCACGAACAACAUGCUCCUAGGCAUCUCCUUCAGGGUGCCCAUGAUGUGCUCGAAGUUGUUCCUGGCU ........--.(((--(.....))))...........(((.((((((((((((....)))))....(((.(((..(.......)..))))))))))))).))).. ( -22.70) >DroAna_CAF1 6239 101 - 1 GGAAAACCAAAGUG-GUAU---UAGAAAAAUAACUUACGACAAAUAACAUGCUUCUCGGCAUUUCUUUCAAAGUGUCCAUGAUAAGUUCAAAAUUUUUCCUAGCU ((((((.....((.-(((.---.............))).)).........((((.(((((((((......)))))))...)).)))).......))))))..... ( -14.64) >consensus UGAUAACA__AGUG_GUAUAUAUAGCUACAAAACUUACCACGAACAACAUGCUUCUGGGCAUCUCCUUCAGGGUGCCCAUGAUGUGCUCGAAGUUGUUCCUGGCU .....................................(((.(((((((..((((.(((((((((......))))))))).))...)).....))))))).))).. (-18.72 = -19.33 + 0.61)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:08:15 2006