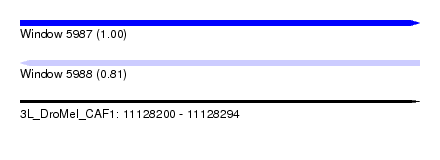
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 11,128,200 – 11,128,294 |
| Length | 94 |
| Max. P | 0.995018 |

| Location | 11,128,200 – 11,128,294 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 73.31 |
| Mean single sequence MFE | -32.53 |
| Consensus MFE | -14.47 |
| Energy contribution | -15.78 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.44 |
| SVM decision value | 2.53 |
| SVM RNA-class probability | 0.995018 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
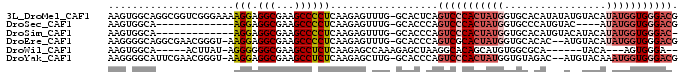
>3L_DroMel_CAF1 11128200 94 + 23771897 AAGUGGCAGGCGGUCGGGAAAAGGAGGCGAAGCCCCUCAAGAGUUUG-GCACUCAGUCCCACUAUGGUGCACAUAUAUGUACAUAUGGUGGGACG .((((.(((((..((..((...((.(((...)))))))..)))))))-.))))..((((((((((((((((......))))).))))))))))). ( -39.80) >DroSec_CAF1 24488 77 + 1 AAGUGGCA-------------AGGAGGCGAAGCCCCUCAAGAGUUUG-GCACCCAGUCCCACUAUGGUGCCCAUGUAC----AUAUGGUGGGACG ..(((.((-------------((((((.(....))))).....))))-.)))...(((((((((((((((....))))----.))))))))))). ( -31.50) >DroSim_CAF1 24397 80 + 1 AAGUGGCA-------------AGGAGGCGAAGCCCCUCAAGAGUUUG-GCACCCAGUCCCACUAUGGUGCACAUGUACAUACAUAUGGUGGGAC- ..(((.((-------------((((((.(....))))).....))))-.)))...(((((((((((((((....)))).....)))))))))))- ( -28.00) >DroEre_CAF1 27417 91 + 1 AAGGGGCAGGCGAACGGGU-AAGGAGGCGAAGCCCCUCAAGAGUUUG-GCACCCAGUCGCACUAUGGUGCACAC--AUGUACAUAUGGUGGGACG ..(((...(.(((((....-.(((.(((...)))))).....)))))-.).))).(((.((((((((((((...--.))))).))))))).))). ( -36.90) >DroWil_CAF1 29184 78 + 1 AAGUGGCA-----ACUUAU-AGGGGGGCGAAGCCUCUCAAGAGCCAAAGAGCUAAGGCACAGCAUGUGGCGCA------UACA---AGUGGGA-- ..(((((.-----......-.(((((((...)))))))....((((....(((.......)))...)))))).------))).---.......-- ( -22.60) >DroYak_CAF1 32413 91 + 1 AAGGGGCAUUCGAACGGGU-AAGGAGGCGAAGCCUCUCAAGAGCUUG-GCACCCAGUCCCACUAUGGUGUAGAC--AUGUACAAAUGGUGGGACG ..(((((((((....))))-..((((((...))))))..........-)).))).((((((((((..((((...--...)))).)))))))))). ( -36.40) >consensus AAGUGGCA_____ACGGGU_AAGGAGGCGAAGCCCCUCAAGAGUUUG_GCACCCAGUCCCACUAUGGUGCACAU__AUGUACAUAUGGUGGGACG .....................(((.(((...))))))..................(((((((((((.................))))))))))). (-14.47 = -15.78 + 1.31)
| Location | 11,128,200 – 11,128,294 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 73.31 |
| Mean single sequence MFE | -25.02 |
| Consensus MFE | -15.47 |
| Energy contribution | -16.67 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.811980 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
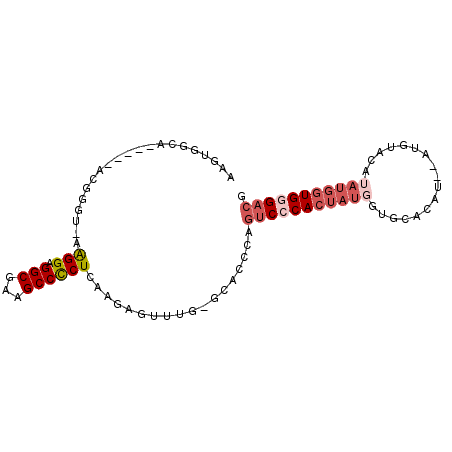
>3L_DroMel_CAF1 11128200 94 - 23771897 CGUCCCACCAUAUGUACAUAUAUGUGCACCAUAGUGGGACUGAGUGC-CAAACUCUUGAGGGGCUUCGCCUCCUUUUCCCGACCGCCUGCCACUU .(((((((.((.((((((....))))))..)).))))))).((((..-...))))..(((((((...))).)))).................... ( -29.10) >DroSec_CAF1 24488 77 - 1 CGUCCCACCAUAU----GUACAUGGGCACCAUAGUGGGACUGGGUGC-CAAACUCUUGAGGGGCUUCGCCUCCU-------------UGCCACUU .(((((((.((.(----((......)))..)).))))))).((((..-...))))..(((((((...))).)))-------------)....... ( -24.00) >DroSim_CAF1 24397 80 - 1 -GUCCCACCAUAUGUAUGUACAUGUGCACCAUAGUGGGACUGGGUGC-CAAACUCUUGAGGGGCUUCGCCUCCU-------------UGCCACUU -((((((((((((((....))))))).......))))))).((((..-...))))..(((((((...))).)))-------------)....... ( -27.31) >DroEre_CAF1 27417 91 - 1 CGUCCCACCAUAUGUACAU--GUGUGCACCAUAGUGCGACUGGGUGC-CAAACUCUUGAGGGGCUUCGCCUCCUU-ACCCGUUCGCCUGCCCCUU .(((.(((.((.(((((..--..)))))..)).))).))).((((((-..(((...((((((((...))).))))-)...))).))..))))... ( -26.30) >DroWil_CAF1 29184 78 - 1 --UCCCACU---UGUA------UGCGCCACAUGCUGUGCCUUAGCUCUUUGGCUCUUGAGAGGCUUCGCCCCCCU-AUAAGU-----UGCCACUU --....(((---((((------...((((...((((.....))))....))))........(((...)))....)-))))))-----........ ( -16.00) >DroYak_CAF1 32413 91 - 1 CGUCCCACCAUUUGUACAU--GUCUACACCAUAGUGGGACUGGGUGC-CAAGCUCUUGAGAGGCUUCGCCUCCUU-ACCCGUUCGAAUGCCCCUU .(((((((....((((...--...)))).....))))))).(((((.-(((....))).(((((...)))))..)-))))............... ( -27.40) >consensus CGUCCCACCAUAUGUACAU__AUGUGCACCAUAGUGGGACUGGGUGC_CAAACUCUUGAGGGGCUUCGCCUCCUU_ACCCGU_____UGCCACUU .(((((((....((((........)))).....))))))).((((......))))....(((((...)))))....................... (-15.47 = -16.67 + 1.20)

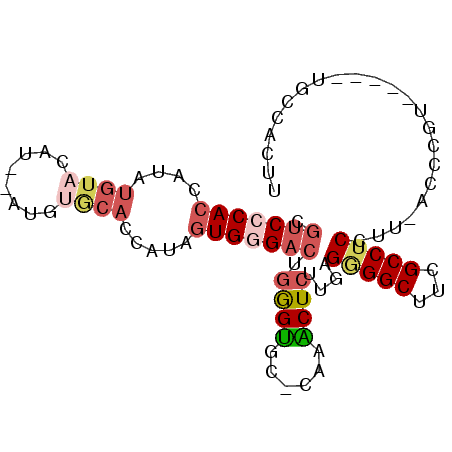
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:08:12 2006