
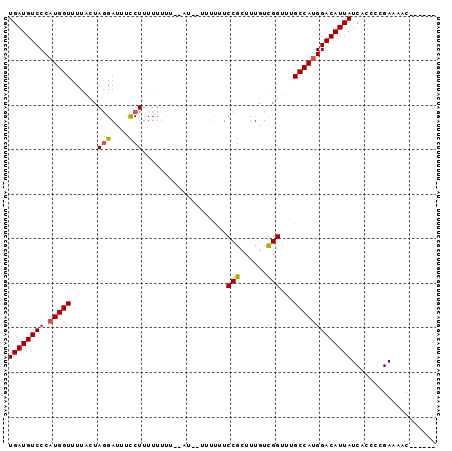
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 11,112,691 – 11,112,781 |
| Length | 90 |
| Max. P | 0.991703 |

| Location | 11,112,691 – 11,112,781 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 81.70 |
| Mean single sequence MFE | -19.80 |
| Consensus MFE | -16.49 |
| Energy contribution | -16.55 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.28 |
| SVM RNA-class probability | 0.991703 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11112691 90 + 23771897 UGAUGUCCCAUGGUUUUCCUAGGAUUUCCUUUUUUUUAUAUCUUUUUUUCCGUUUUGUCGGUUUGCCAUGGACAUUAUCGCCCCGAAAAC------ ((((((((.(((((......(((....)))...................(((......)))...))))))))))))).............------ ( -20.20) >DroSec_CAF1 9468 88 + 1 UGAUGUCCCAUGGUUUUACUAGGAUUUCCUUUUUUUUCCAU--UUUUUUCCGCUUUGUUGGUUUGCCAUGGACAUUAUCACCCCGAAAAC------ ((((((((.(((((...(((((((............)))..--........((...))))))..))))))))))))).............------ ( -18.40) >DroSim_CAF1 9195 78 + 1 UGAUGUCCCAUGGUUUUACUAGGAUUUCCUUUUU------------UUUCCGCUUUGUUGGUUUGCCAUGGACAUUAUCACCCCGAAAAC------ ((((((((.(((((...(((((((..........------------..))).......))))..))))))))))))).............------ ( -18.81) >DroEre_CAF1 11708 94 + 1 UGAUGUCCCAUGGUUUUCUUAAGAUUUUCUGUUUUCUUGUU--UUUCUUCCGUUUUGUCGGCUUGCCAGGGACAUUAUCGCCCCGAAAACCGAAAA (((((((((.((((.....(((((..........)))))..--......(((......)))...)))))))))))))................... ( -21.80) >consensus UGAUGUCCCAUGGUUUUACUAGGAUUUCCUUUUUUUU__AU__UUUUUUCCGCUUUGUCGGUUUGCCAUGGACAUUAUCACCCCGAAAAC______ ((((((((.(((((......(((....)))...................(((......)))...)))))))))))))................... (-16.49 = -16.55 + 0.06)


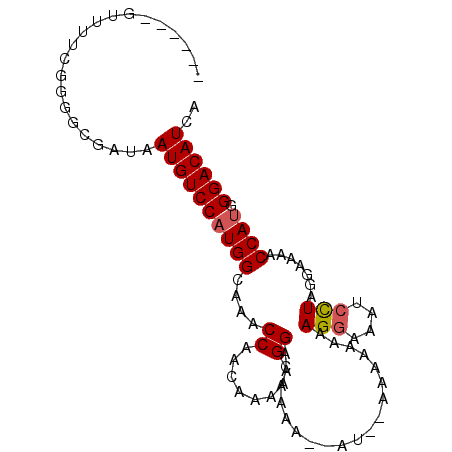
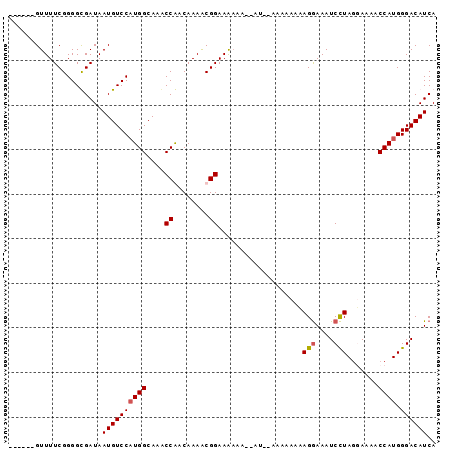
| Location | 11,112,691 – 11,112,781 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 81.70 |
| Mean single sequence MFE | -17.32 |
| Consensus MFE | -12.64 |
| Energy contribution | -12.95 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.708952 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11112691 90 - 23771897 ------GUUUUCGGGGCGAUAAUGUCCAUGGCAAACCGACAAAACGGAAAAAAAGAUAUAAAAAAAAGGAAAUCCUAGGAAAACCAUGGGACAUCA ------.((((((((((......)))).(((....)))......))))))..................((..(((((((....)).)))))..)). ( -17.70) >DroSec_CAF1 9468 88 - 1 ------GUUUUCGGGGUGAUAAUGUCCAUGGCAAACCAACAAAGCGGAAAAAA--AUGGAAAAAAAAGGAAAUCCUAGUAAAACCAUGGGACAUCA ------...............((((((((((....((........))......--...........(((....))).......)))).)))))).. ( -15.30) >DroSim_CAF1 9195 78 - 1 ------GUUUUCGGGGUGAUAAUGUCCAUGGCAAACCAACAAAGCGGAAA------------AAAAAGGAAAUCCUAGUAAAACCAUGGGACAUCA ------...............((((((((((....((........))...------------....(((....))).......)))).)))))).. ( -15.30) >DroEre_CAF1 11708 94 - 1 UUUUCGGUUUUCGGGGCGAUAAUGUCCCUGGCAAGCCGACAAAACGGAAGAAA--AACAAGAAAACAGAAAAUCUUAAGAAAACCAUGGGACAUCA (((((.(((((((((((......))))).......(((......)))......--.....)))))).)))))(((((.(.....).)))))..... ( -21.00) >consensus ______GUUUUCGGGGCGAUAAUGUCCAUGGCAAACCAACAAAACGGAAAAAA__AU__AAAAAAAAGGAAAUCCUAGGAAAACCAUGGGACAUCA .....................((((((((((....((........))...................(((....))).......)))).)))))).. (-12.64 = -12.95 + 0.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:08:10 2006