
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 11,098,238 – 11,098,370 |
| Length | 132 |
| Max. P | 0.872881 |

| Location | 11,098,238 – 11,098,330 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 85.36 |
| Mean single sequence MFE | -19.98 |
| Consensus MFE | -13.44 |
| Energy contribution | -13.00 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.814825 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11098238 92 + 23771897 UCCAGUGACUCACAAGCUGGCUUCAAAUAAUGAAGCCUAACUUAAGGCGUCAGUUGCAUCCUUGCAAAUUUUUGUAAAUUCAUUUAAUAAGC ...(((((...((((((((((((((.....))))((((......))))))))))((((....)))).....))))....)))))........ ( -20.80) >DroSec_CAF1 9251 87 + 1 UCCAGUGACUCACGAGCUGGCUUCGAAAAAUGAAGCCUAACUUAAGGCGUCACUUGCAUCCUUAAAUUUUGUUGGAAAUUCAUUUAG----- (((((((((.(..(((..(((((((.....)))))))...)))...).)))))..(((...........)))))))...........----- ( -20.80) >DroSim_CAF1 11674 87 + 1 UCCAGUGACUCACAAGCUGGCUUCAAAAAAUGAAGCCUAACUUAAGGCGUCAGUUGCAUCCUUGAAUUUUGUUGUAAAUUCAUUUAA----- ....(..(((.((..((((((((((.....)))))))........))))).)))..).....(((((((......))))))).....----- ( -21.00) >DroEre_CAF1 9454 92 + 1 UCCAGUUACUCACUAGCUGGCUUCAAAAAAUGAAGCCUAACUUAAGGCGUCAUUUGCAUCCUUGAAUAUUUUAGUAAAUACUUUUAGUAAAC .(((((((.....))))))).(((((.((((((.((((......)))).))))))......)))))...((((.((((....)))).)))). ( -21.00) >DroYak_CAF1 9416 92 + 1 UCCAGUUAUUCACAAGCUGGCUUUAAAAAAUGAAGCCUAACUUAAGGCGCCAUUUGCAUCCUUGAAUUUUUUACUUAAUACUUUUAAUAACC ....((((((...(((..(((((((.....)))))))....((((((.((.....))..))))))...............)))..)))))). ( -16.30) >consensus UCCAGUGACUCACAAGCUGGCUUCAAAAAAUGAAGCCUAACUUAAGGCGUCAGUUGCAUCCUUGAAUUUUUUUGUAAAUUCAUUUAAUAA_C ..................(((((((.....)))))))....((((((.((.....))..))))))........................... (-13.44 = -13.00 + -0.44)

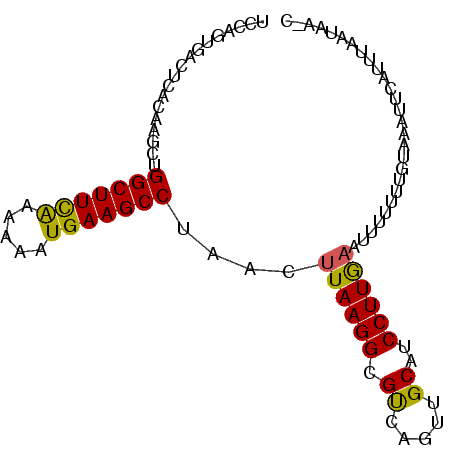

| Location | 11,098,238 – 11,098,330 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 85.36 |
| Mean single sequence MFE | -23.00 |
| Consensus MFE | -15.68 |
| Energy contribution | -16.92 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.872881 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11098238 92 - 23771897 GCUUAUUAAAUGAAUUUACAAAAAUUUGCAAGGAUGCAACUGACGCCUUAAGUUAGGCUUCAUUAUUUGAAGCCAGCUUGUGAGUCACUGGA .........................(((((....))))).((((....((((((.(((((((.....)))))))))))))...))))..... ( -23.70) >DroSec_CAF1 9251 87 - 1 -----CUAAAUGAAUUUCCAACAAAAUUUAAGGAUGCAAGUGACGCCUUAAGUUAGGCUUCAUUUUUCGAAGCCAGCUCGUGAGUCACUGGA -----.....(((((((......)))))))........((((((..(...((((.((((((.......))))))))))...).))))))... ( -21.60) >DroSim_CAF1 11674 87 - 1 -----UUAAAUGAAUUUACAACAAAAUUCAAGGAUGCAACUGACGCCUUAAGUUAGGCUUCAUUUUUUGAAGCCAGCUUGUGAGUCACUGGA -----.....(((((((......)))))))..........((((....((((((.(((((((.....)))))))))))))...))))..... ( -24.30) >DroEre_CAF1 9454 92 - 1 GUUUACUAAAAGUAUUUACUAAAAUAUUCAAGGAUGCAAAUGACGCCUUAAGUUAGGCUUCAUUUUUUGAAGCCAGCUAGUGAGUAACUGGA .....(((....(((((((((........((((...(....)...))))..(((.(((((((.....)))))))))))))))))))..))). ( -24.10) >DroYak_CAF1 9416 92 - 1 GGUUAUUAAAAGUAUUAAGUAAAAAAUUCAAGGAUGCAAAUGGCGCCUUAAGUUAGGCUUCAUUUUUUAAAGCCAGCUUGUGAAUAACUGGA (((((((..((((...........((((.((((.(((.....))))))).)))).(((((.........))))).))))...)))))))... ( -21.30) >consensus G_UUAUUAAAUGAAUUUACAAAAAAAUUCAAGGAUGCAAAUGACGCCUUAAGUUAGGCUUCAUUUUUUGAAGCCAGCUUGUGAGUCACUGGA .............((((((..........((((...(....)...))))(((((.(((((((.....))))))))))))))))))....... (-15.68 = -16.92 + 1.24)

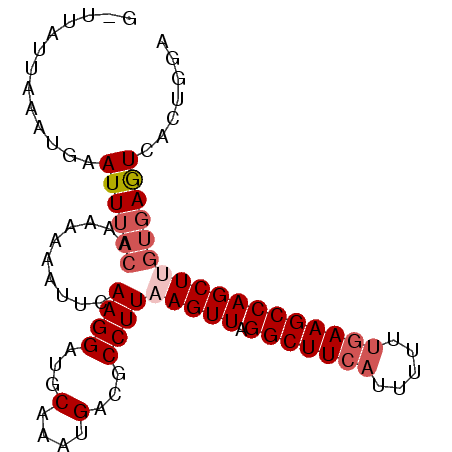
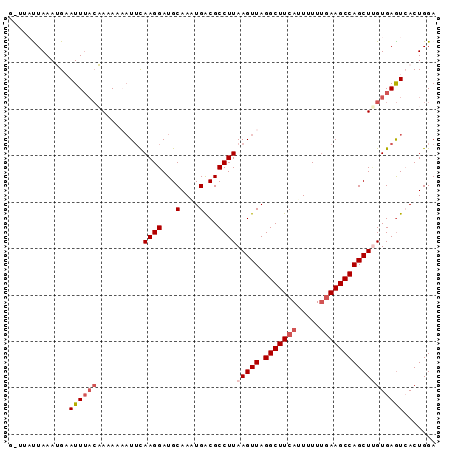
| Location | 11,098,250 – 11,098,370 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.05 |
| Mean single sequence MFE | -22.38 |
| Consensus MFE | -14.84 |
| Energy contribution | -14.16 |
| Covariance contribution | -0.68 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.702254 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11098250 120 + 23771897 CAAGCUGGCUUCAAAUAAUGAAGCCUAACUUAAGGCGUCAGUUGCAUCCUUGCAAAUUUUUGUAAAUUCAUUUAAUAAGCGCUCUAUGGCCCAACAUAUUCCAGUUUCCAAAAGUUUUUA .((((((((((.....((((((((((......)))).....(((((....)))))...........))))))....))))....((((......))))...))))))............. ( -25.00) >DroSec_CAF1 9263 109 + 1 CGAGCUGGCUUCGAAAAAUGAAGCCUAACUUAAGGCGUCACUUGCAUCCUUAAAUUUUGUUGGAAAUUCAUUUAG-----------UGACACAACAUAUUCCAGUUUCCAAAAGUUUUUA .((((((((((((.....)))))))....((((((.((.....))..))))))......(((((((((....(((-----------(......)).))....))))))))).)))))... ( -22.20) >DroSim_CAF1 11686 109 + 1 CAAGCUGGCUUCAAAAAAUGAAGCCUAACUUAAGGCGUCAGUUGCAUCCUUGAAUUUUGUUGUAAAUUCAUUUAA-----------UGUCCCAACAUAUUCCAGUUUCCAAAAGUUUUUA .(((((((..........(((.((((......)))).)))(((((((...(((((((......)))))))....)-----------))...)))).....)))))))............. ( -22.60) >DroEre_CAF1 9466 108 + 1 CUAGCUGGCUUCAAAAAAUGAAGCCUAACUUAAGGCGUCAUUUGCAUCCUUGAAUAUUUUAGUAAAUACUUUUAGUAAACGCCUCAUGAUUCAACAUAUUCUAAUUAC------------ ......(((((((.....))))))).......((((((((..........)))....((((.((((....)))).)))))))))........................------------ ( -19.60) >DroYak_CAF1 9428 120 + 1 CAAGCUGGCUUUAAAAAAUGAAGCCUAACUUAAGGCGCCAUUUGCAUCCUUGAAUUUUUUACUUAAUACUUUUAAUAACCGCUUUAUGGUCCAAUAUAUGCCAGUUUCUAAAAGUUUGUA .((((((((.............((((......))))(((((..((....(((((................))))).....))...))))).........))))))))............. ( -22.49) >consensus CAAGCUGGCUUCAAAAAAUGAAGCCUAACUUAAGGCGUCAGUUGCAUCCUUGAAUUUUUUUGUAAAUUCAUUUAAUAA_CGC___AUGACCCAACAUAUUCCAGUUUCCAAAAGUUUUUA .((((((((((((.....)))))))....((((((.((.....))..)))))).................................................)))))............. (-14.84 = -14.16 + -0.68)

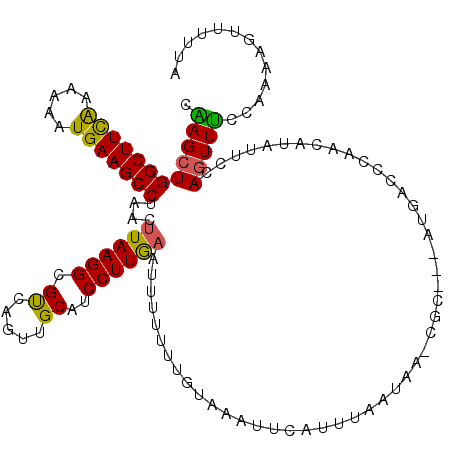
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:08:07 2006