
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 11,095,458 – 11,095,600 |
| Length | 142 |
| Max. P | 0.999204 |

| Location | 11,095,458 – 11,095,560 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 74.02 |
| Mean single sequence MFE | -25.80 |
| Consensus MFE | -21.25 |
| Energy contribution | -21.03 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.968802 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11095458 102 + 23771897 GAAA-------------CAACCAGUUAUGCCAAUAUAAUUUUUUUUUUUUGUAUGAUCCCAUAGUGCGUCCCAACGAAGGCGAUGCCUAUCCAUUGGGCGCCUUGCCCGUGGGCA ....-------------..........((((.((((((..........))))))......((((.(((((((......)).))))))))).(((.((((.....))))))))))) ( -25.70) >DroEre_CAF1 6626 109 + 1 GAAAUACACUGUAACACUAAAAUAUUA--AAAAUA---UUCAU-UAUUUUGUAUAUUCCCGAAGUGCGUCCCAACGAAGGCGAUGCCUAUCCACUGGGCGCUCUGCCCGUGGGCA ((((((((..((((......(((((..--...)))---))..)-)))..))))).)))....((.(((((((......)).))))))).(((((.((((.....))))))))).. ( -26.40) >DroYak_CAF1 6594 97 + 1 ---------------ACUAAAAUAUUA--AACAUAUAAUUCAU-UAUUUUGUGUAUUCCCAAAGUGCGACCCAACGAAGGCGAUGCCUAUCCAUUGGGCGCCCUGCCCGUGGGCA ---------------............--.(((((.(((....-.))).)))))........((.(((.(((((.((((((...)))).))..)))))))).))(((....))). ( -25.30) >consensus GAAA___________ACUAAAAUAUUA__AAAAUAUAAUUCAU_UAUUUUGUAUAUUCCCAAAGUGCGUCCCAACGAAGGCGAUGCCUAUCCAUUGGGCGCCCUGCCCGUGGGCA ..............................................................((.(((.((((..((((((...)))).))...))))))).))(((....))). (-21.25 = -21.03 + -0.22)



| Location | 11,095,458 – 11,095,560 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 74.02 |
| Mean single sequence MFE | -30.93 |
| Consensus MFE | -25.99 |
| Energy contribution | -25.77 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.84 |
| SVM decision value | 3.43 |
| SVM RNA-class probability | 0.999204 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11095458 102 - 23771897 UGCCCACGGGCAAGGCGCCCAAUGGAUAGGCAUCGCCUUCGUUGGGACGCACUAUGGGAUCAUACAAAAAAAAAAAAUUAUAUUGGCAUAACUGGUUG-------------UUUC ..((((..(.....(((((((((((..((((...)))))))))))).))).)..))))................((((...((..(.....)..)).)-------------))). ( -28.50) >DroEre_CAF1 6626 109 - 1 UGCCCACGGGCAGAGCGCCCAGUGGAUAGGCAUCGCCUUCGUUGGGACGCACUUCGGGAAUAUACAAAAUA-AUGAA---UAUUUU--UAAUAUUUUAGUGUUACAGUGUAUUUC ((((....))))..(((((((((((..((((...)))))))))))).)))......(((((((((..((((-..(((---(((...--..))))))...))))...))))))))) ( -33.40) >DroYak_CAF1 6594 97 - 1 UGCCCACGGGCAGGGCGCCCAAUGGAUAGGCAUCGCCUUCGUUGGGUCGCACUUUGGGAAUACACAAAAUA-AUGAAUUAUAUGUU--UAAUAUUUUAGU--------------- ..((((.((.....(((((((((((..((((...)))))))))))).))).)).)))).......((((((-.(((((.....)))--)).))))))...--------------- ( -30.90) >consensus UGCCCACGGGCAGGGCGCCCAAUGGAUAGGCAUCGCCUUCGUUGGGACGCACUUUGGGAAUAUACAAAAUA_AUGAAUUAUAUUUU__UAAUAUUUUAGU___________UUUC ((((....))))..(((((((((((..((((...)))))))))))).)))................................................................. (-25.99 = -25.77 + -0.22)

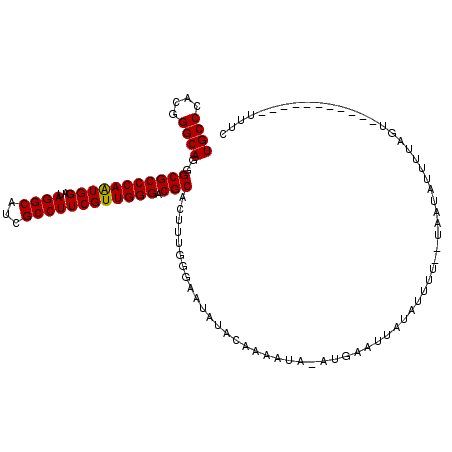
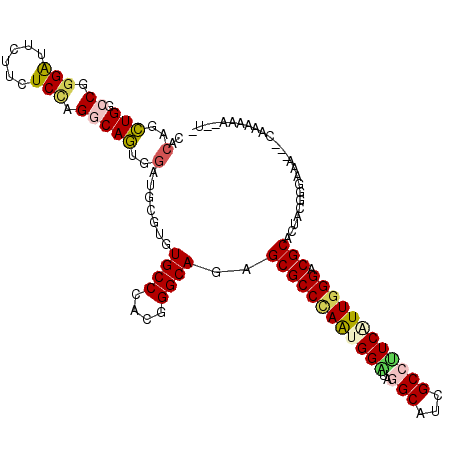
| Location | 11,095,485 – 11,095,600 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 77.78 |
| Mean single sequence MFE | -40.95 |
| Consensus MFE | -27.51 |
| Energy contribution | -27.85 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.731335 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
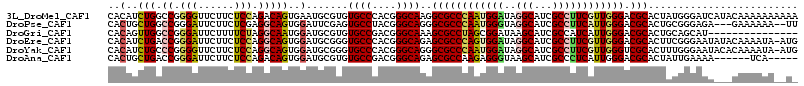
>3L_DroMel_CAF1 11095485 115 - 23771897 CACAUCUGGCCGGGGUUCUUCUCCAGACAGUGAAUGCGUGUGCCCACGGGCAAGGCGCCCAAUGGAUAGGCAUCGCCUUCGUUGGGACGCACUAUGGGAUCAUACAAAAAAAAAA ..((.(((.(.((((.....)))).).))))).....(((((((((..(.....(((((((((((..((((...)))))))))))).))).)..))))..))))).......... ( -40.60) >DroPse_CAF1 10726 110 - 1 CACUGCUGGCCGGGAUUCUUCUCGAGGCAGUGGAUUCGAGUGCCUACGGGCAGGGCGCCCAAUGGGUAGGCAUCGCCUUCAUUGGGACGCACUGCGGGAGA---GAAAAAA--UU (((((((...(((((....))))).)))))))..((((((((((....)))...(((((((((((..((((...)))))))))))).)))))).))))...---.......--.. ( -48.50) >DroGri_CAF1 7009 100 - 1 CACAGUUGGCCGGGAUUCUUUUCUAGGCAAUGGAUGCGUGUGCCGACGGGCAAAGCGCCUAGCGGAUAAGCAUCGCCAUCAUUGGGACGCACUGCAGCAU--------------- ....(((((((.(((......))).)))....(((((.(((.(((..((((.....))))..)))))).))))).(((....))).........))))..--------------- ( -33.80) >DroEre_CAF1 6661 114 - 1 CACAUCUGACCGGGAUUCUUCUCCAGGCAGUGGAUGCGGGUGCCCACGGGCAGAGCGCCCAGUGGAUAGGCAUCGCCUUCGUUGGGACGCACUUCGGGAAUAUACAAAAUA-AUG ..((.(((.((.(((......))).)))))))..(((((((((((((((((.....)))).)))....)))))).(((.....))).))))(....)..............-... ( -36.50) >DroYak_CAF1 6617 114 - 1 CACAUCUGCCCGGGGUUCUUCUCCAGGCAGUGGAUGCGGGUGCCCACGGGCAGGGCGCCCAAUGGAUAGGCAUCGCCUUCGUUGGGUCGCACUUUGGGAAUACACAAAAUA-AUG ....(((((((((((((((.(((((.....)))).).))).)))).))))))))(((((((((((..((((...)))))))))))).)))..((((........))))...-... ( -48.60) >DroAna_CAF1 6886 104 - 1 CACUGCUGACCGGGAUUCUUCUCCAGACAGUGGAUGCGUGUGCCGACGGGCAGAGCGCCAAGAGGGUAAGCAUCGCCCUCAUUGGGACGCACUAUUGAAAA------UCA----- ((((((((...((((....))))))).)))))..(((((.((((....)))).....((((((((((.......)))))).)))).)))))..........------...----- ( -37.70) >consensus CACAGCUGGCCGGGAUUCUUCUCCAGGCAGUGGAUGCGUGUGCCCACGGGCAGAGCGCCCAAUGGAUAGGCAUCGCCUUCAUUGGGACGCACUACGGGAAA___CAAAAAA__U_ ..(..(((.((.(((......))).)))))..).......((((....))))..((((((((((((..(((...)))))))))))).)))......................... (-27.51 = -27.85 + 0.34)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:08:04 2006