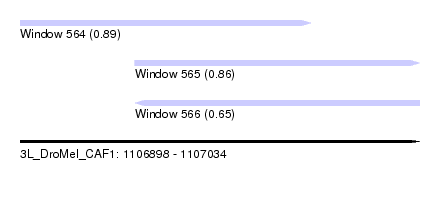
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,106,898 – 1,107,034 |
| Length | 136 |
| Max. P | 0.894974 |

| Location | 1,106,898 – 1,106,997 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 88.00 |
| Mean single sequence MFE | -30.37 |
| Consensus MFE | -28.70 |
| Energy contribution | -28.70 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.894974 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1106898 99 + 23771897 ACUCUACACACUCUACACACUCCAAUUCGGAGGCCGUCCGUCAAUGUCCAUGUCCUGCUGCUGGACAUUGGCGAAAAAUGCGUUAGCCAACGUUGCCGG------ ...................((((.....)))).(((..((((((((((((.((......))))))))))))))......(((((....)))))...)))------ ( -29.60) >DroSec_CAF1 390 90 + 1 ACUC---------UACACACUCCAAUUCGGAGGCCGUCCGUCAAUGUCCAUGUCCUGCUGCUGGACAUUGGCGAAAAAUGCGUUAGCCAACGUUGCCGC------ ....---------......((((.....))))......((((((((((((.((......))))))))))))))......(((.((((....)))).)))------ ( -29.00) >DroYak_CAF1 229 96 + 1 UCUC---------CACACACUCCAAUUCGGAGGCCGUCCGUCAAUGUCCAUGUCCUGCUGCUGGACAUUGGCGAAAAAUGCGUUAGCCAACGUUGCCGGUUGCCG ....---------..............(((..((((..((((((((((((.((......))))))))))))))......(((((....)))))...))))..))) ( -32.50) >consensus ACUC_________UACACACUCCAAUUCGGAGGCCGUCCGUCAAUGUCCAUGUCCUGCUGCUGGACAUUGGCGAAAAAUGCGUUAGCCAACGUUGCCGG______ ....................(((.....)))(((....((((((((((((.((......))))))))))))))......(((((....))))).)))........ (-28.70 = -28.70 + -0.00)

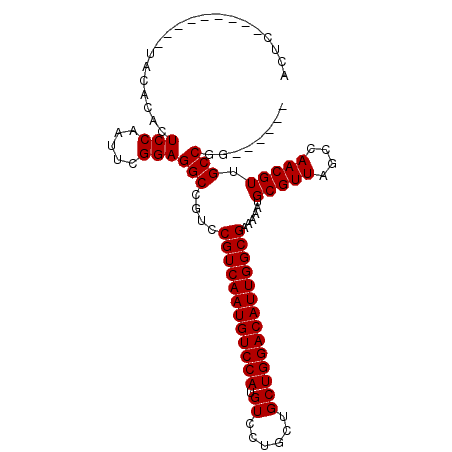
| Location | 1,106,937 – 1,107,034 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.33 |
| Mean single sequence MFE | -39.42 |
| Consensus MFE | -22.19 |
| Energy contribution | -21.82 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.858944 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1106937 97 + 23771897 GUCAAUGUCC-------------AUGUCCUGCUGCUGGACAUUGGCGAAAAAUGCGUUAGCCA-ACGUUGCCGG-------UU--GCCGGCUUCCUCGCCGAGGAAAAGUAGAAGGCAAA ((((((((((-------------(.((......))))))))))))).............(((.-.....(((((-------..--.)))))((((((...))))))........)))... ( -36.90) >DroPse_CAF1 490 118 + 1 GUCAAUGUCCAUGUCCAGUGUCCAUGUCCUGGUGCUGGACAUUGGCGAAAAAUGCGUUAGCCAAACGACGCCGC-CGCCA-UCGAGGAGUCGUCCGGGACUCUGGAAAGUAGAAGGCAAA (((((((((((.((((((.(.......))))).))))))))))))).............(((...((((.((..-((...-.)).)).))))(((((....)))))........)))... ( -42.40) >DroSec_CAF1 420 97 + 1 GUCAAUGUCC-------------AUGUCCUGCUGCUGGACAUUGGCGAAAAAUGCGUUAGCCA-ACGUUGCCGC-------UU--GCCGACUUCCUCGCCGAGGAAAAGUAGAAGGCAAA ((((((((((-------------(.((......))))))))))))).......(((.((((..-..)))).)))-------((--(((.((((((((...)))))..)))....))))). ( -35.90) >DroYak_CAF1 259 104 + 1 GUCAAUGUCC-------------AUGUCCUGCUGCUGGACAUUGGCGAAAAAUGCGUUAGCCA-ACGUUGCCGGUUGCCGGUU--GCCGGCUUCCUCGCCGAGCAAAAGUAGAAGGCAAA ((((((((((-------------(.((......))))))))))))).......(((((....)-))))((((..((((...((--((((((......)))).))))..))))..)))).. ( -42.50) >consensus GUCAAUGUCC_____________AUGUCCUGCUGCUGGACAUUGGCGAAAAAUGCGUUAGCCA_ACGUUGCCGC_______UU__GCCGGCUUCCUCGCCGAGGAAAAGUAGAAGGCAAA ((((((((((..........................)))))))))).............(((...((....))..............((((......)))).............)))... (-22.19 = -21.82 + -0.37)

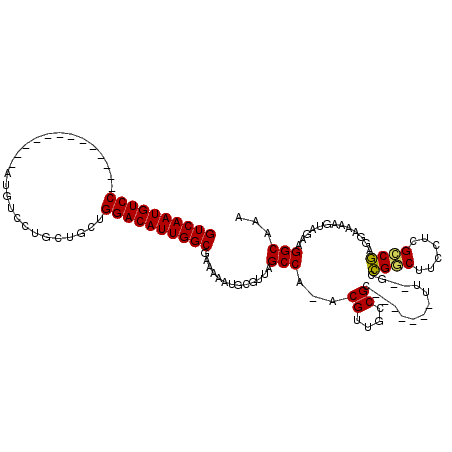
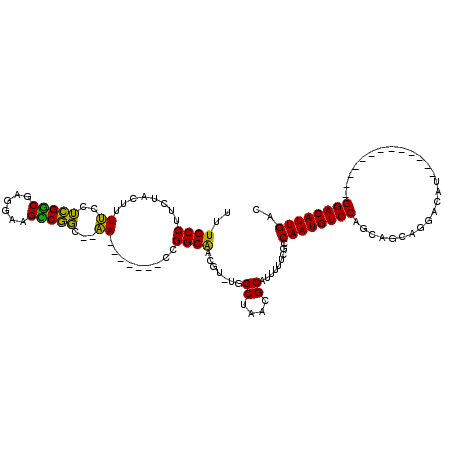
| Location | 1,106,937 – 1,107,034 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.33 |
| Mean single sequence MFE | -35.85 |
| Consensus MFE | -23.61 |
| Energy contribution | -21.80 |
| Covariance contribution | -1.81 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.652894 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1106937 97 - 23771897 UUUGCCUUCUACUUUUCCUCGGCGAGGAAGCCGGC--AA-------CCGGCAACGU-UGGCUAACGCAUUUUUCGCCAAUGUCCAGCAGCAGGACAU-------------GGACAUUGAC ...............(((..((((((((((((((.--..-------))))).....-..((....)).))))))))).((((((.......))))))-------------)))....... ( -34.00) >DroPse_CAF1 490 118 - 1 UUUGCCUUCUACUUUCCAGAGUCCCGGACGACUCCUCGA-UGGCG-GCGGCGUCGUUUGGCUAACGCAUUUUUCGCCAAUGUCCAGCACCAGGACAUGGACACUGGACAUGGACAUUGAC .......((((...(((((.((((((((((((.(((((.-...))-).)).))))))))((....))...........((((((.......)))))))))).)))))..))))....... ( -40.50) >DroSec_CAF1 420 97 - 1 UUUGCCUUCUACUUUUCCUCGGCGAGGAAGUCGGC--AA-------GCGGCAACGU-UGGCUAACGCAUUUUUCGCCAAUGUCCAGCAGCAGGACAU-------------GGACAUUGAC ...............(((..(((((((((((..((--.(-------(((....)))-).))..))...))))))))).((((((.......))))))-------------)))....... ( -33.20) >DroYak_CAF1 259 104 - 1 UUUGCCUUCUACUUUUGCUCGGCGAGGAAGCCGGC--AACCGGCAACCGGCAACGU-UGGCUAACGCAUUUUUCGCCAAUGUCCAGCAGCAGGACAU-------------GGACAUUGAC ....................((((((((((((((.--..)))))...((....)).-..((....)).)))))))))((((((((.(.....)...)-------------)))))))... ( -35.70) >consensus UUUGCCUUCUACUUUUCCUCGGCGAGGAAGCCGGC__AA_______CCGGCAACGU_UGGCUAACGCAUUUUUCGCCAAUGUCCAGCAGCAGGACAU_____________GGACAUUGAC ..((((........((..(((((......)))))...)).........)))).......((....)).........((((((((..........................)))))))).. (-23.61 = -21.80 + -1.81)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:40:37 2006