
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 11,059,873 – 11,059,972 |
| Length | 99 |
| Max. P | 0.895442 |

| Location | 11,059,873 – 11,059,972 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 89.97 |
| Mean single sequence MFE | -28.80 |
| Consensus MFE | -22.89 |
| Energy contribution | -22.75 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.16 |
| Mean z-score | -3.12 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.895442 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
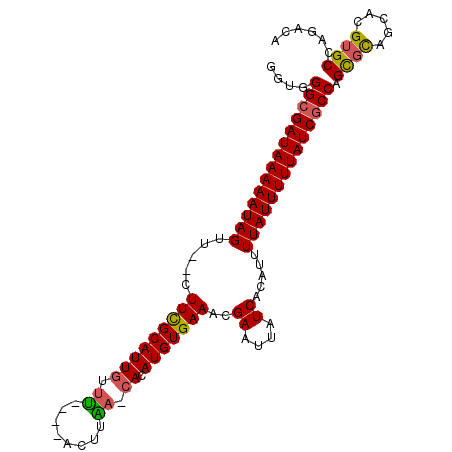
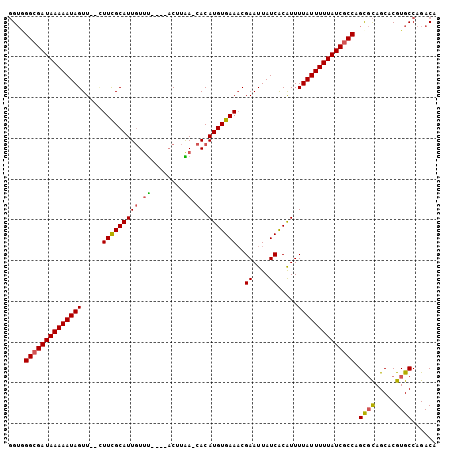
>3L_DroMel_CAF1 11059873 99 + 23771897 GGUGGGCGAUAAAAAUAGUU--CUUCGCAUUGUUU----ACUUAACCACAUGUGAAACGAAUUAUCACAUUUUAUUUUUAUCGCCAGCGCAGCACGUGCCAGACA ....((((((((((((((..--.(((((((.(((.----....)))...)))))))..((....)).....)))))))))))))).((((.....))))...... ( -26.90) >DroSec_CAF1 3875 98 + 1 GGUGGGCGAUAAAAAUAGUU--CUUCGCAUUGUGU----ACUUAA-CACAUGUGAAACGAAUUAUCACAUUUUAUUUUUAUCGCCAGCGCAGCACGUGCCAGACA ....((((((((((((((..--.(((((((.((((----.....)-))))))))))..((....)).....)))))))))))))).((((.....))))...... ( -31.70) >DroSim_CAF1 3809 98 + 1 GGUGGGCGAUAAAAAUAGUU--CUUCGCAUUGUUU----ACUUAA-CACAUGUGAAACGAAUUAUCACAUUUUAUUUUUAUCGCCAGCGCAGCACGUGCCAGACA ....((((((((((((((..--.(((((((((((.----....))-)).)))))))..((....)).....)))))))))))))).((((.....))))...... ( -29.50) >DroEre_CAF1 3879 98 + 1 GGUAGGCGAUAAAAAUAGUU--UUUCGCAUUGUUU----ACUUGA-CACAUGUGAAACGAAUUAUCACAUUUUAUUUUUAUCGCCAGUGCAGCACGUGCCAGACU ((((((((((((((((((..--((((((((((((.----....))-)).)))))))).((....)).....)))))))))))))).(((...))).))))..... ( -31.50) >DroYak_CAF1 3733 97 + 1 GGUAGGCGAUAAAAAUAGUU---UUCGCAUUGUUU----ACUUGA-CACAUGUGAAACGAAUUAUCACAUUUUAUUUUUAUCGCCAGCGCAGCACGUGCCAGACA ....((((((((((((((.(---(((((((((((.----....))-)).)))))))).((....)).....)))))))))))))).((((.....))))...... ( -30.80) >DroAna_CAF1 3464 104 + 1 CGAUGGCGAUAAAAAUAGUUGUUUUUGCAUUCUUCUGCCCCCCGA-CACAUGUGAAUCGAAUUAUCGCGUUUUAUUUUUAUCACCAGCCUAGCACGUGCCAGACA ...(((.((((((((((((((.....(((......)))....)))-)..((((((.........))))))..)))))))))).)))(.((.((....)).)).). ( -22.40) >consensus GGUGGGCGAUAAAAAUAGUU__CUUCGCAUUGUUU____ACUUAA_CACAUGUGAAACGAAUUAUCACAUUUUAUUUUUAUCGCCAGCGCAGCACGUGCCAGACA ....((((((((((((((.....(((((((((.((........)).)).)))))))..((....)).....)))))))))))))).((((.....))))...... (-22.89 = -22.75 + -0.14)

| Location | 11,059,873 – 11,059,972 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 89.97 |
| Mean single sequence MFE | -27.66 |
| Consensus MFE | -21.71 |
| Energy contribution | -21.63 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.629350 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
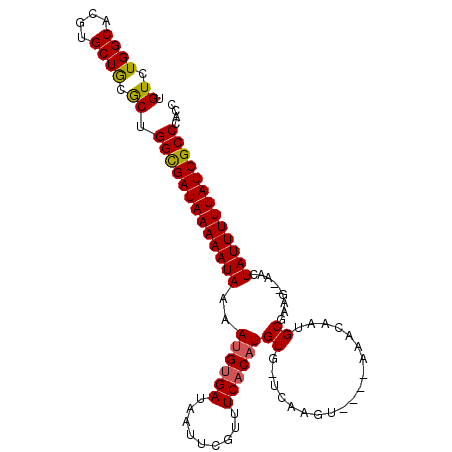
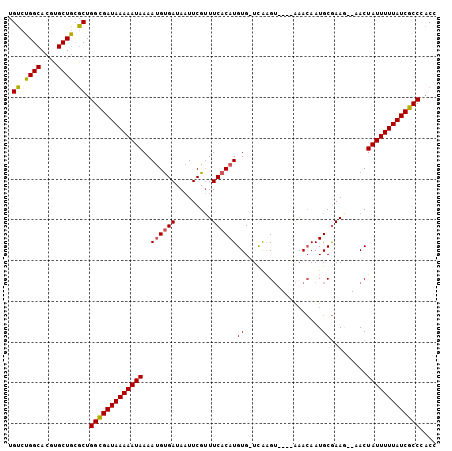
>3L_DroMel_CAF1 11059873 99 - 23771897 UGUCUGGCACGUGCUGCGCUGGCGAUAAAAAUAAAAUGUGAUAAUUCGUUUCACAUGUGGUUAAGU----AAACAAUGCGAAG--AACUAUUUUUAUCGCCCACC .((.((((....)))).)).(((((((((((((..((((((.........))))))...(((..((----(.....)))....--)))))))))))))))).... ( -26.00) >DroSec_CAF1 3875 98 - 1 UGUCUGGCACGUGCUGCGCUGGCGAUAAAAAUAAAAUGUGAUAAUUCGUUUCACAUGUG-UUAAGU----ACACAAUGCGAAG--AACUAUUUUUAUCGCCCACC .((.((((....)))).)).(((((((((((((..((((((.........))))))(((-(.....----)))).........--...))))))))))))).... ( -28.40) >DroSim_CAF1 3809 98 - 1 UGUCUGGCACGUGCUGCGCUGGCGAUAAAAAUAAAAUGUGAUAAUUCGUUUCACAUGUG-UUAAGU----AAACAAUGCGAAG--AACUAUUUUUAUCGCCCACC .((.((((....)))).)).(((((((((((((..((((((.........))))))..(-((..((----(.....)))....--)))))))))))))))).... ( -26.00) >DroEre_CAF1 3879 98 - 1 AGUCUGGCACGUGCUGCACUGGCGAUAAAAAUAAAAUGUGAUAAUUCGUUUCACAUGUG-UCAAGU----AAACAAUGCGAAA--AACUAUUUUUAUCGCCUACC (((.((((....)))).)))(((((((((((((..((((((.........))))))((.-....((----(.....)))....--.))))))))))))))).... ( -26.80) >DroYak_CAF1 3733 97 - 1 UGUCUGGCACGUGCUGCGCUGGCGAUAAAAAUAAAAUGUGAUAAUUCGUUUCACAUGUG-UCAAGU----AAACAAUGCGAA---AACUAUUUUUAUCGCCUACC .((.((((....)))).)).(((((((((((((..((((((.........))))))((.-....((----(.....)))...---.))))))))))))))).... ( -25.60) >DroAna_CAF1 3464 104 - 1 UGUCUGGCACGUGCUAGGCUGGUGAUAAAAAUAAAACGCGAUAAUUCGAUUCACAUGUG-UCGGGGGGCAGAAGAAUGCAAAAACAACUAUUUUUAUCGCCAUCG .(((((((....)))))))((((((((((((((...(.(((((..............))-))).)..(((......))).........))))))))))))))... ( -33.14) >consensus UGUCUGGCACGUGCUGCGCUGGCGAUAAAAAUAAAAUGUGAUAAUUCGUUUCACAUGUG_UCAAGU____AAACAAUGCGAAG__AACUAUUUUUAUCGCCCACC .((.((((....)))).)).(((((((((((((..((((((.........))))))((...................)).........))))))))))))).... (-21.71 = -21.63 + -0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:07:45 2006