
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 11,010,548 – 11,010,688 |
| Length | 140 |
| Max. P | 0.997294 |

| Location | 11,010,548 – 11,010,656 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 73.02 |
| Mean single sequence MFE | -21.87 |
| Consensus MFE | -12.47 |
| Energy contribution | -12.19 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.57 |
| SVM decision value | 2.19 |
| SVM RNA-class probability | 0.990066 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
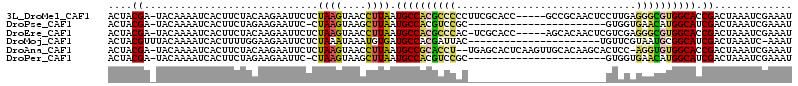
>3L_DroMel_CAF1 11010548 108 + 23771897 ACUACGA-UACAAAAUCACUUCUACAAGAAUUCUCUAAGUAACCUUAAUGCCACGCCCCCUUCGCACC-----GCCGCAACUCCUUGAGGGCGUGGCACCGACUAAAUCGAAAU ....(((-(........((((.....(((....)))))))........((((((((((.....((...-----...))..(.....).))))))))))........)))).... ( -25.20) >DroPse_CAF1 155895 89 + 1 ACUACGA-UACAAAAUCACUUCUAGAAGAAUUC-CUAAGUAAGCUUAAUGCCACGUCCGC-----------------------GUGGUGAACAUGGCAUCGACUAAAUCGAAAU ..(((((-(.....))).(((....))).....-....))).(((....((((((....)-----------------------)))))......))).((((.....))))... ( -16.40) >DroEre_CAF1 144419 107 + 1 ACUACGA-UACAAAAUCACUUCUACAAGAAUUCUCUAAGUAACCUUAAUGCCACGCCCAC-UCGCACC-----AGCACAACUCGUCGAGGGCGUGGCACCGACUAAAUCGAAAU ....(((-(........((((.....(((....)))))))........((((((((((..-(((.((.-----((.....)).)))))))))))))))........)))).... ( -28.10) >DroMoj_CAF1 160406 91 + 1 ACUACGUUUACAAAAUCACUUUUGGAAGAAUUCUCUAAAUAAAUGUGAUGCCACGAUUAC----------------------UGUUCGUAAUGCGGCAUCGACUAAAUC-AAAU ...((((((((((((....)))))..(((....)))...)))))))((((((.(.(((((----------------------.....)))))).)))))).........-.... ( -19.40) >DroAna_CAF1 137642 110 + 1 ACUACGA-UACAAAAUCACUUCUACAAGAAUUCUCUAAGUAACCUUAAUGCCGCACCU--UGAGCACUCAAGUUGCACAAGCACUCC-AGGUGUGGCACCGACUAAAUCGAAAU ....(((-(........((((.....(((....)))))))........((((((((((--.(((.........(((....)))))).-))))))))))........)))).... ( -25.70) >DroPer_CAF1 142231 89 + 1 ACUACGA-UACAAAAUCACUUCUAGAAGAAUUC-CUAAGUAAGCUUAAUGCCACGUCCGC-----------------------GUGGUGAACAUGGCAUCGACUAAAUCGAAAU ..(((((-(.....))).(((....))).....-....))).(((....((((((....)-----------------------)))))......))).((((.....))))... ( -16.40) >consensus ACUACGA_UACAAAAUCACUUCUACAAGAAUUCUCUAAGUAACCUUAAUGCCACGCCCAC_______________________GUCGAGAACGUGGCACCGACUAAAUCGAAAU ....((.............................((((....)))).((((((((((..............................)))))))))).))............. (-12.47 = -12.19 + -0.27)


| Location | 11,010,548 – 11,010,656 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 73.02 |
| Mean single sequence MFE | -32.47 |
| Consensus MFE | -20.67 |
| Energy contribution | -19.64 |
| Covariance contribution | -1.02 |
| Combinations/Pair | 1.37 |
| Mean z-score | -3.08 |
| Structure conservation index | 0.64 |
| SVM decision value | 2.83 |
| SVM RNA-class probability | 0.997294 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
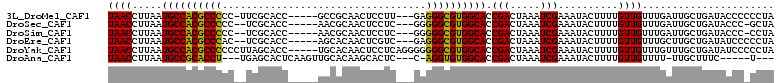
>3L_DroMel_CAF1 11010548 108 - 23771897 AUUUCGAUUUAGUCGGUGCCACGCCCUCAAGGAGUUGCGGC-----GGUGCGAAGGGGGCGUGGCAUUAAGGUUACUUAGAGAAUUCUUGUAGAAGUGAUUUUGUA-UCGUAGU ....((((......((((((((((((((......(((((..-----..))))).))))))))))))))((((((((((.(((....)))....))))))))))..)-))).... ( -39.90) >DroPse_CAF1 155895 89 - 1 AUUUCGAUUUAGUCGAUGCCAUGUUCACCAC-----------------------GCGGACGUGGCAUUAAGCUUACUUAG-GAAUUCUUCUAGAAGUGAUUUUGUA-UCGUAGU ....((((..((((((((((((((((.....-----------------------..))))))))))))..((((...(((-((....))))).))))))))....)-))).... ( -26.40) >DroEre_CAF1 144419 107 - 1 AUUUCGAUUUAGUCGGUGCCACGCCCUCGACGAGUUGUGCU-----GGUGCGA-GUGGGCGUGGCAUUAAGGUUACUUAGAGAAUUCUUGUAGAAGUGAUUUUGUA-UCGUAGU ....((((......(((((((((((((((((.((.....))-----.)).)))-..))))))))))))((((((((((.(((....)))....))))))))))..)-))).... ( -40.40) >DroMoj_CAF1 160406 91 - 1 AUUU-GAUUUAGUCGAUGCCGCAUUACGAACA----------------------GUAAUCGUGGCAUCACAUUUAUUUAGAGAAUUCUUCCAAAAGUGAUUUUGUAAACGUAGU ....-......((.(((((((((((((.....----------------------))))).)))))))))).....((((.(((((((((....))).)))))).))))...... ( -23.30) >DroAna_CAF1 137642 110 - 1 AUUUCGAUUUAGUCGGUGCCACACCU-GGAGUGCUUGUGCAACUUGAGUGCUCA--AGGUGCGGCAUUAAGGUUACUUAGAGAAUUCUUGUAGAAGUGAUUUUGUA-UCGUAGU ....((((......((((((.(((((-.(((..((((.......))))..))).--))))).))))))((((((((((.(((....)))....))))))))))..)-))).... ( -38.40) >DroPer_CAF1 142231 89 - 1 AUUUCGAUUUAGUCGAUGCCAUGUUCACCAC-----------------------GCGGACGUGGCAUUAAGCUUACUUAG-GAAUUCUUCUAGAAGUGAUUUUGUA-UCGUAGU ....((((..((((((((((((((((.....-----------------------..))))))))))))..((((...(((-((....))))).))))))))....)-))).... ( -26.40) >consensus AUUUCGAUUUAGUCGAUGCCACGCCCACAAC_______________________GCGGGCGUGGCAUUAAGGUUACUUAGAGAAUUCUUCUAGAAGUGAUUUUGUA_UCGUAGU ..............((((((((((((..............................))))))))))))....((((.((.(((((((((....))).)))))).))...)))). (-20.67 = -19.64 + -1.02)



| Location | 11,010,586 – 11,010,688 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 82.54 |
| Mean single sequence MFE | -28.67 |
| Consensus MFE | -21.12 |
| Energy contribution | -20.43 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.882888 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11010586 102 + 23771897 UAACCUUAAUGCCACGCCCCC-UUCGCACC-----GCCGCAACUCCUU---GAGGGCGUGGCACCGACUAAAUCGAAAUACUUUUGUUGUUUGAUUGCUGAUACCCCCCUA .........((((((((((..-...((...-----...))..(.....---).)))))))))).......(((((((..((....))..)))))))............... ( -26.30) >DroSec_CAF1 147080 100 + 1 UAACCUUAAUGCCACGCCCCC--UCGCACC-----AACGCAACUCCUC---GGGGGCGUGGCACCGACUAAAUCGAAAUACUUUUGUUGUUUGAUUGCUGAUACCC-GCUA .........((((((((((((--.......-----.............---)))))))))))).......(((((((..((....))..)))))))((........-)).. ( -31.55) >DroSim_CAF1 142184 100 + 1 UAACCUUAAUGCCACGCCCCC--UCGCACC-----AACGCAACUCCUC---GGGGGCGUGGCACCGACUAAAUCGAAAUACUUUUGUUGUUUGAUUGCUGAUACCC-CCUA .........((((((((((((--.......-----.............---)))))))))))).......(((((((..((....))..)))))))..........-.... ( -31.15) >DroEre_CAF1 144457 101 + 1 UAACCUUAAUGCCACGCCCAC--UCGCACC-----AGCACAACUCGUC---GAGGGCGUGGCACCGACUAAAUCGAAAUACUUUUGUUGUUUGCUUGCUGAUAUCCCCCUA ((((.....((((((((((..--(((.((.-----((.....)).)))---)))))))))))).(((.....)))..........))))...................... ( -25.70) >DroYak_CAF1 147640 106 + 1 UAACCUUAAUGCCACGCCCCCCUUAGCACC-----UGCACAACUCCUCAGGGGGGGCGUGGCACCGACUAAAUCGAAAUACUUUUGUUGUUUGUUUGCUGAUAUCCCCCUA ((((.....(((((((((((((...((...-----.))....((....))))))))))))))).(((.....)))..........))))...................... ( -33.30) >DroAna_CAF1 137680 95 + 1 UAACCUUAAUGCCGCACCU---UGAGCACUCAAGUUGCACAAGCACUC---C-AGGUGUGGCACCGACUAAAUCGAAAUACUUUUGUUGUUUU-UUGCUUUC-----U--- .........((((((((((---.(((.........(((....))))))---.-))))))))))..........(((((.((.......)).))-))).....-----.--- ( -24.00) >consensus UAACCUUAAUGCCACGCCCCC__UCGCACC_____AGCACAACUCCUC___GGGGGCGUGGCACCGACUAAAUCGAAAUACUUUUGUUGUUUGAUUGCUGAUACCC_CCUA ((((.....((((((((((..................................)))))))))).(((.....)))..........))))...................... (-21.12 = -20.43 + -0.69)
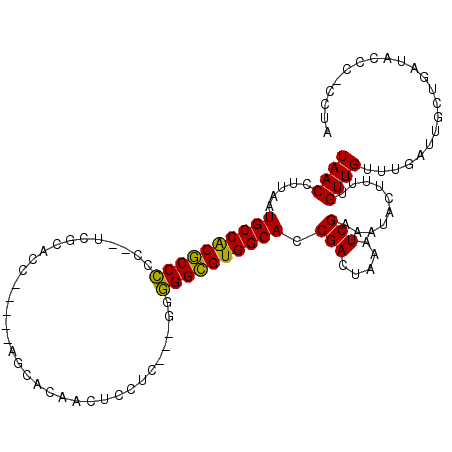

| Location | 11,010,586 – 11,010,688 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 82.54 |
| Mean single sequence MFE | -33.41 |
| Consensus MFE | -27.01 |
| Energy contribution | -27.17 |
| Covariance contribution | 0.15 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.829446 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 11010586 102 - 23771897 UAGGGGGGUAUCAGCAAUCAAACAACAAAAGUAUUUCGAUUUAGUCGGUGCCACGCCCUC---AAGGAGUUGCGGC-----GGUGCGAA-GGGGGCGUGGCAUUAAGGUUA ....(..((((...................))))..).........((((((((((((((---......(((((..-----..))))).-))))))))))))))....... ( -31.71) >DroSec_CAF1 147080 100 - 1 UAGC-GGGUAUCAGCAAUCAAACAACAAAAGUAUUUCGAUUUAGUCGGUGCCACGCCCCC---GAGGAGUUGCGUU-----GGUGCGA--GGGGGCGUGGCAUUAAGGUUA ..((-........))((((.................((((...))))(((((((((((((---..(.(.(......-----).).)..--)))))))))))))...)))). ( -35.90) >DroSim_CAF1 142184 100 - 1 UAGG-GGGUAUCAGCAAUCAAACAACAAAAGUAUUUCGAUUUAGUCGGUGCCACGCCCCC---GAGGAGUUGCGUU-----GGUGCGA--GGGGGCGUGGCAUUAAGGUUA ...(-..((((...................))))..).........((((((((((((((---..(.(.(......-----).).)..--))))))))))))))....... ( -34.31) >DroEre_CAF1 144457 101 - 1 UAGGGGGAUAUCAGCAAGCAAACAACAAAAGUAUUUCGAUUUAGUCGGUGCCACGCCCUC---GACGAGUUGUGCU-----GGUGCGA--GUGGGCGUGGCAUUAAGGUUA ....(..((((...................))))..).........((((((((((((((---(((.((.....))-----.)).)))--..))))))))))))....... ( -31.91) >DroYak_CAF1 147640 106 - 1 UAGGGGGAUAUCAGCAAACAAACAACAAAAGUAUUUCGAUUUAGUCGGUGCCACGCCCCCCCUGAGGAGUUGUGCA-----GGUGCUAAGGGGGGCGUGGCAUUAAGGUUA ....(..((((...................))))..).........(((((((((((((((...((.(.((....)-----).).))..)))))))))))))))....... ( -37.61) >DroAna_CAF1 137680 95 - 1 ---A-----GAAAGCAA-AAAACAACAAAAGUAUUUCGAUUUAGUCGGUGCCACACCU-G---GAGUGCUUGUGCAACUUGAGUGCUCA---AGGUGCGGCAUUAAGGUUA ---.-----........-.....(((..........((((...))))(((((.(((((-.---(((..((((.......))))..))).---))))).)))))....))). ( -29.00) >consensus UAGG_GGGUAUCAGCAAUCAAACAACAAAAGUAUUUCGAUUUAGUCGGUGCCACGCCCCC___GAGGAGUUGCGCU_____GGUGCGA__GGGGGCGUGGCAUUAAGGUUA .......................(((..........((((...))))(((((((((((((.........((((((.......))))))..)))))))))))))....))). (-27.01 = -27.17 + 0.15)

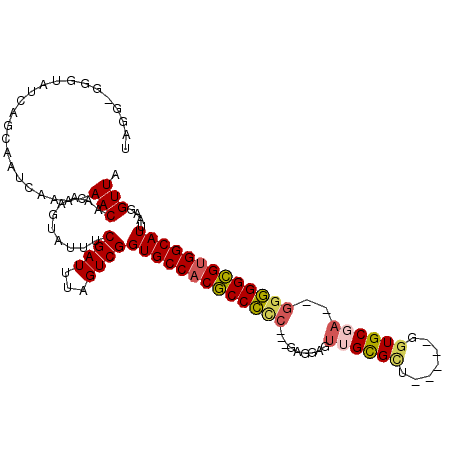

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:07:23 2006