| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,103,004 – 1,103,109 |
| Length | 105 |
| Max. P | 0.965735 |

| Location | 1,103,004 – 1,103,109 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.61 |
| Mean single sequence MFE | -25.18 |
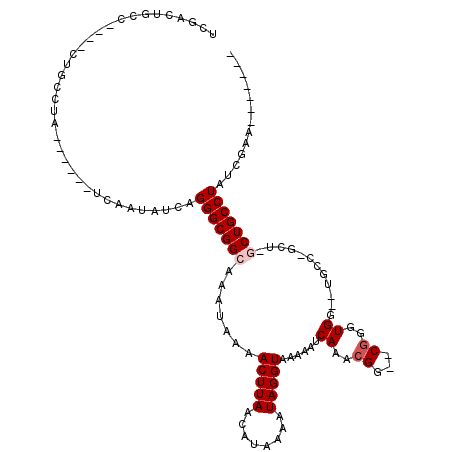
| Consensus MFE | -11.54 |
| Energy contribution | -12.14 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.46 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.965735 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1103004 105 + 23771897 UCGACUGCC----CUGCCUAUCAAUAUCAAUAUCAGGGCGGCAAAUAAAACUUAACAUAAAAUAGGUAAAAAUCAAACGG--CGGGUGG--UGCCAGCUAGCUGCCUAUCGAA------- (((((((((----(((..(((........))).))))))))........(((((........)))))...........((--(((.(((--(....)))).)))))..)))).------- ( -30.30) >DroSec_CAF1 20275 85 + 1 -------------------A------UCAAUAUCAGGGCGGCAAAUAAAACUUAACAUAAAAUAGGUAAAAAUCAAACGC--C-GGUGGUUUGCCUGCUCGCUGCCUAUCGAA------- -------------------.------.........(((((((.......(((((........)))))...........((--.-(((.....))).))..)))))))......------- ( -20.40) >DroSim_CAF1 13426 101 + 1 UCGACUGCC----CUGCCUA------UCAAUAUCAGGGCGGCAAAUAAAACUUAACAUAAAAUAGGUAAAAAUCAAACGG--CGGGUGGUUUGCCUGCUCGCUGCCUAUCGAA------- ((((..(((----(((....------.......))))))(((.......(((((........)))))...........((--(((((((....)).))))))))))..)))).------- ( -31.70) >DroEre_CAF1 14316 84 + 1 --------------UGCCUA------UCAAUAUCAGGGCGGCAAAUAAAACUUAACAUAAAAUAGGUAAAAAUCAAACGGGGCGGGUGG--UGC-------CUGCCUAUCGAA------- --------------.((((.------.........))))..........(((((........)))))..........(((((((((...--..)-------))))))..))..------- ( -18.80) >DroYak_CAF1 16153 104 + 1 UCGACUGCCCGCCCCGCCUA------UCAAUAUCAGGGCGGCAAAUAAAACUUAACAUAAAAUAGGUAAAAAUCAAACGGAGCGGCUGC---CU-------CUGCCUAUCGAACUCAAAC ((((..(((((..((((((.------.........))))))........(((((........)))))..........))).))(((...---..-------..)))..))))........ ( -24.70) >consensus UCGACUGCC____CUGCCUA______UCAAUAUCAGGGCGGCAAAUAAAACUUAACAUAAAAUAGGUAAAAAUCAAACGG__CGGGUGG__UGCC_GCU_GCUGCCUAUCGAA_______ ...................................(((((((.......(((((........)))))......((..((...))..))............)))))))............. (-11.54 = -12.14 + 0.60)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:40:32 2006