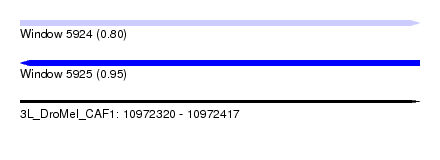
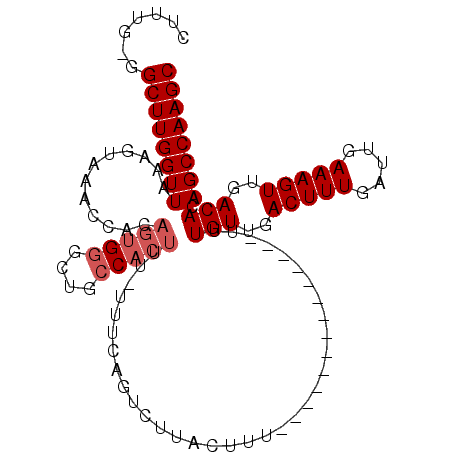
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,972,320 – 10,972,417 |
| Length | 97 |
| Max. P | 0.945393 |

| Location | 10,972,320 – 10,972,417 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 85.44 |
| Mean single sequence MFE | -23.00 |
| Consensus MFE | -17.72 |
| Energy contribution | -19.38 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.802824 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10972320 97 + 23771897 GCUUGGCUGUGUCAACUUUCAAUCAAAGUCAACAAAAAAA----------AAAGUAAGACUGAAA-AAAGUGGCAACCCACUGUGGUUUACUUUAACCAAGCC-CAAAG ((((((...(((..(((((.....)))))..)))......----------((((((.(((((...-..(((((....))))).)))))))))))..)))))).-..... ( -24.00) >DroSec_CAF1 104463 92 + 1 GCUUGGCUGUGUCAACUUUCAAUCAAAGUCAACAAA--------------AAAGUAAGACUGAAA--AAGUGGCAGCCCACUGUGGUUUACUUUAACCAAGCC-CAAAG ((((((...(((..(((((.....)))))..)))..--------------((((((.(((((...--.(((((....))))).)))))))))))..)))))).-..... ( -24.70) >DroSim_CAF1 102674 93 + 1 GCUUGGCUGUGUCAACUUUCAAUCAAAGUCAACAAA--------------AAAGUAAGACUGAAA-AAAGUGGCAGCCCACUGUGGUUUACUUUAACCAAGCC-CAAAG ((((((...(((..(((((.....)))))..)))..--------------((((((.(((((...-..(((((....))))).)))))))))))..)))))).-..... ( -24.00) >DroEre_CAF1 105132 94 + 1 GCUUGGCUGUGUCAACUUUCAAUCAAAGUCAACAAA--------------AAAGUAAGACUGAAA-AAAGUGGCAGCCCACUGUGGUUUACUUUAACCAAGCCCCCAAG ((((((...(((..(((((.....)))))..)))..--------------((((((.(((((...-..(((((....))))).)))))))))))..))))))....... ( -24.00) >DroYak_CAF1 107821 108 + 1 GCUUGGCUGUGUCAACUUUCAAUCAAAGUCAACAAAAAAAAAAAUGGAAAAAAGUAAGACUGAAAAAAAGUGGCAAACCACUGUGGUUUACUUUAACCAAGCC-CAAAG ((((((...(((..(((((.....)))))..)))................((((((.(((((......(((((....))))).)))))))))))..)))))).-..... ( -23.70) >DroAna_CAF1 98489 74 + 1 GCUUGGCUGUGUCAACUUUCAAUCAAAGUCAACAAA--------------AAAGGA---------------G---GCACACUGUGGUUUACUUUAACCAAGCC-C--AG .((.(((((((((..((((.................--------------))))..---------------)---))))....(((((......)))))))))-.--)) ( -17.63) >consensus GCUUGGCUGUGUCAACUUUCAAUCAAAGUCAACAAA______________AAAGUAAGACUGAAA_AAAGUGGCAGCCCACUGUGGUUUACUUUAACCAAGCC_CAAAG ((((((...(((..(((((.....)))))..)))................((((((.(((((......(((((....))))).)))))))))))..))))))....... (-17.72 = -19.38 + 1.67)

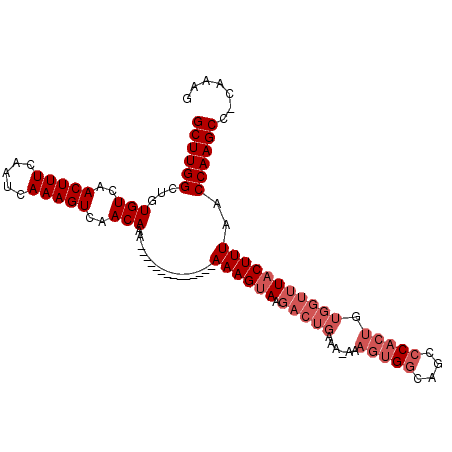
| Location | 10,972,320 – 10,972,417 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 85.44 |
| Mean single sequence MFE | -26.03 |
| Consensus MFE | -22.35 |
| Energy contribution | -23.02 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.945393 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10972320 97 - 23771897 CUUUG-GGCUUGGUUAAAGUAAACCACAGUGGGUUGCCACUUU-UUUCAGUCUUACUUU----------UUUUUUUGUUGACUUUGAUUGAAAGUUGACACAGCCAAGC .....-.(((((((((((((((((...(((((....)))))..-.....)).)))))))----------......(((..(((((.....)))))..))).)))))))) ( -27.10) >DroSec_CAF1 104463 92 - 1 CUUUG-GGCUUGGUUAAAGUAAACCACAGUGGGCUGCCACUU--UUUCAGUCUUACUUU--------------UUUGUUGACUUUGAUUGAAAGUUGACACAGCCAAGC .....-.(((((((((((((((((...(((((....))))).--.....)).)))))))--------------..(((..(((((.....)))))..))).)))))))) ( -27.70) >DroSim_CAF1 102674 93 - 1 CUUUG-GGCUUGGUUAAAGUAAACCACAGUGGGCUGCCACUUU-UUUCAGUCUUACUUU--------------UUUGUUGACUUUGAUUGAAAGUUGACACAGCCAAGC .....-.(((((((((((((((((...(((((....)))))..-.....)).)))))))--------------..(((..(((((.....)))))..))).)))))))) ( -27.10) >DroEre_CAF1 105132 94 - 1 CUUGGGGGCUUGGUUAAAGUAAACCACAGUGGGCUGCCACUUU-UUUCAGUCUUACUUU--------------UUUGUUGACUUUGAUUGAAAGUUGACACAGCCAAGC .......(((((((((((((((((...(((((....)))))..-.....)).)))))))--------------..(((..(((((.....)))))..))).)))))))) ( -27.10) >DroYak_CAF1 107821 108 - 1 CUUUG-GGCUUGGUUAAAGUAAACCACAGUGGUUUGCCACUUUUUUUCAGUCUUACUUUUUUCCAUUUUUUUUUUUGUUGACUUUGAUUGAAAGUUGACACAGCCAAGC .....-.((((((((...(((((((.....)))))))......................................(((..(((((.....)))))..))).)))))))) ( -27.40) >DroAna_CAF1 98489 74 - 1 CU--G-GGCUUGGUUAAAGUAAACCACAGUGUGC---C---------------UCCUUU--------------UUUGUUGACUUUGAUUGAAAGUUGACACAGCCAAGC ..--.-.((((((((...(((.((....)).)))---.---------------......--------------..(((..(((((.....)))))..))).)))))))) ( -19.80) >consensus CUUUG_GGCUUGGUUAAAGUAAACCACAGUGGGCUGCCACUUU_UUUCAGUCUUACUUU______________UUUGUUGACUUUGAUUGAAAGUUGACACAGCCAAGC .......((((((((............(((((....)))))..................................(((..(((((.....)))))..))).)))))))) (-22.35 = -23.02 + 0.67)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:06:58 2006