| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,964,823 – 10,964,920 |
| Length | 97 |
| Max. P | 0.988495 |

| Location | 10,964,823 – 10,964,920 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 77.83 |
| Mean single sequence MFE | -36.68 |
| Consensus MFE | -15.65 |
| Energy contribution | -16.68 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.39 |
| Structure conservation index | 0.43 |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.988495 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
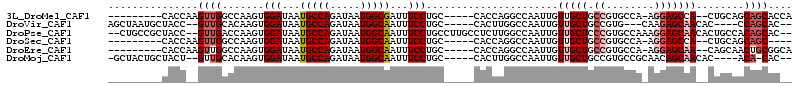
>3L_DroMel_CAF1 10964823 97 + 23771897 ---------CACCAAGUUGGCCAAGUGGAUAAUGCCAGAUAAUGGCGAUUUCCUGC-----CACCAGGCCAAUUGUUGCUGCCGUGCCA-AGGAGCCA--CUGCAGCAGCACCA ---------.....((((((((..((((..(((((((.....)))).))).....)-----)))..))))))))((((((((.(((.(.-....).))--).)))))))).... ( -41.20) >DroVir_CAF1 88479 98 + 1 AGCUAAUGCUACC--GUUGCACAAGUGGAUAAUGCCAGAUAAUGGCAAUUUCCUGC-----CACUUGGCCAAUUGUUGCUGCCGUG---CAAGAGCAACAC----CCAGCAC-- ......((((...--(((((.(((((((....(((((.....)))))........)-----))))))......(.((((......)---))).))))))..----..)))).-- ( -30.90) >DroPse_CAF1 101554 108 + 1 --CUGCCGCUACC--GUUGACCAAGUGGAUAAUGCCAGAUAAUGGCAAUUUCCUGCCUUGCCUCUUGGCCAAUUGUUGCUCCCGUGCCAAAGGAGCAACACUGCCACAGCAC-- --...........--((((..((((.(((...(((((.....)))))...)))...)))).....((((....(((((((((.........)))))))))..))))))))..-- ( -35.80) >DroSec_CAF1 97073 93 + 1 ---------CACCAAGUUGGCCAAGUGGAUAAUGCCAGAUAAUGGCAAUUUCCUGC-----CACCAGGCCAAUUGUUGCUGCCGUGCCA-AGGAGCCA--CUGCAGCAGC---- ---------.....((((((((..((((....(((((.....)))))........)-----)))..))))))))((((((((.(((.(.-....).))--).))))))))---- ( -39.60) >DroEre_CAF1 97732 97 + 1 ---------CACCAAGUUGGCCAAGUGGAUAAUGCCAGAUAAUGGCAAUUUCCUGC-----CACCAGGCCAAUUGUUGCUGCCGUGCCA-AGGAGCAA--CAGCAACUGCGGCA ---------..((.((((((((..((((....(((((.....)))))........)-----)))..))))))))(((((((...(((..-....))).--)))))))...)).. ( -38.40) >DroMoj_CAF1 97379 99 + 1 -GCUACUGCUACU--GUUGCACAAGUGGAUAAUGCCAGAUAAUGGCAAUUUCCUGC-----CACUUGGCCAAUUGUUGCUGCCGUGCCGCAACAGCAACAC----ACA-CAC-- -((....))...(--(((((.(((((((....(((((.....)))))........)-----))))))......((((((.((...)).)))))))))))).----...-...-- ( -34.20) >consensus _________CACC__GUUGGCCAAGUGGAUAAUGCCAGAUAAUGGCAAUUUCCUGC_____CACCAGGCCAAUUGUUGCUGCCGUGCCA_AGGAGCAA__CUGCAACAGCAC__ ...............((((.......(((...(((((.....)))))...)))......................(((((.((........)))))))........)))).... (-15.65 = -16.68 + 1.03)
| Location | 10,964,823 – 10,964,920 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 77.83 |
| Mean single sequence MFE | -40.65 |
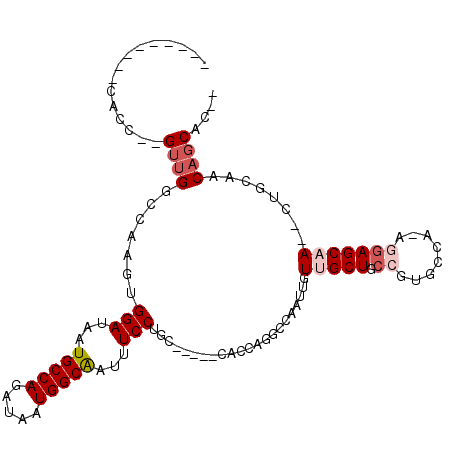
| Consensus MFE | -18.04 |
| Energy contribution | -18.98 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.27 |
| Structure conservation index | 0.44 |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.987551 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10964823 97 - 23771897 UGGUGCUGCUGCAG--UGGCUCCU-UGGCACGGCAGCAACAAUUGGCCUGGUG-----GCAGGAAAUCGCCAUUAUCUGGCAUUAUCCACUUGGCCAACUUGGUG--------- ......((((((.(--((.(....-.).))).)))))).(((((((((.((((-----(...(....)((((.....)))).....))))).)))))).)))...--------- ( -39.20) >DroVir_CAF1 88479 98 - 1 --GUGCUGG----GUGUUGCUCUUG---CACGGCAGCAACAAUUGGCCAAGUG-----GCAGGAAAUUGCCAUUAUCUGGCAUUAUCCACUUGUGCAAC--GGUAGCAUUAGCU --..(((((----.(((((((.(((---((((((...........)))(((((-----(........(((((.....)))))....)))))))))))).--)))))))))))). ( -38.20) >DroPse_CAF1 101554 108 - 1 --GUGCUGUGGCAGUGUUGCUCCUUUGGCACGGGAGCAACAAUUGGCCAAGAGGCAAGGCAGGAAAUUGCCAUUAUCUGGCAUUAUCCACUUGGUCAAC--GGUAGCGGCAG-- --.((((((.((..((((((((((.......)))))))))).(((((((((..((...)).(((...(((((.....)))))...))).))))))))).--.)).)))))).-- ( -47.80) >DroSec_CAF1 97073 93 - 1 ----GCUGCUGCAG--UGGCUCCU-UGGCACGGCAGCAACAAUUGGCCUGGUG-----GCAGGAAAUUGCCAUUAUCUGGCAUUAUCCACUUGGCCAACUUGGUG--------- ----..((((((.(--((.(....-.).))).)))))).(((((((((.((((-----(........(((((.....)))))....))))).)))))).)))...--------- ( -40.50) >DroEre_CAF1 97732 97 - 1 UGCCGCAGUUGCUG--UUGCUCCU-UGGCACGGCAGCAACAAUUGGCCUGGUG-----GCAGGAAAUUGCCAUUAUCUGGCAUUAUCCACUUGGCCAACUUGGUG--------- .(((...(((((((--(((..(..-..)..))))))))))..((((((.((((-----(........(((((.....)))))....))))).))))))...))).--------- ( -41.50) >DroMoj_CAF1 97379 99 - 1 --GUG-UGU----GUGUUGCUGUUGCGGCACGGCAGCAACAAUUGGCCAAGUG-----GCAGGAAAUUGCCAUUAUCUGGCAUUAUCCACUUGUGCAAC--AGUAGCAGUAGC- --...-...----.((((((((((((.((......)).((((...(((....)-----)).(((...(((((.....)))))...)))..)))))))))--))))))).....- ( -36.70) >consensus __GUGCUGUUGCAG__UUGCUCCU_UGGCACGGCAGCAACAAUUGGCCAAGUG_____GCAGGAAAUUGCCAUUAUCUGGCAUUAUCCACUUGGCCAAC__GGUA_________ ................(((((((........)).)))))...((((((.(((.........(((...(((((.....)))))...)))))).))))))................ (-18.04 = -18.98 + 0.94)

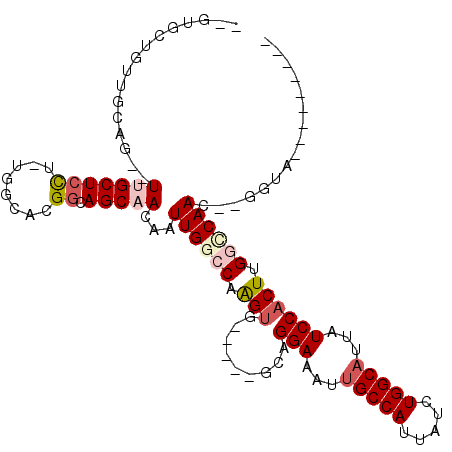
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:06:56 2006