
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,948,589 – 10,948,726 |
| Length | 137 |
| Max. P | 0.956687 |

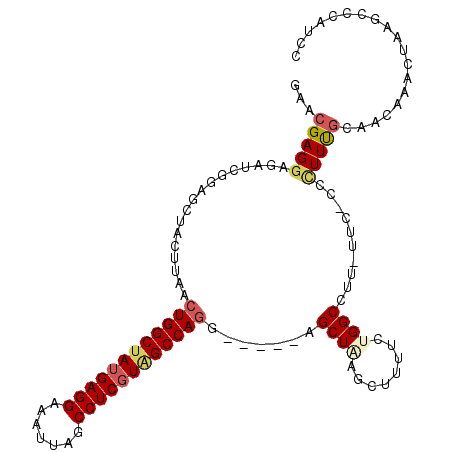
| Location | 10,948,589 – 10,948,696 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 82.90 |
| Mean single sequence MFE | -35.41 |
| Consensus MFE | -22.52 |
| Energy contribution | -23.22 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.607032 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10948589 107 + 23771897 GAACGAGGAGAUCGGAGCUACUUAACUGGCUAUGAGGAAAUUAGCCUCGUAGCCAGG-----AGCUCAGCUUUUCUGGCCUUUUGC-CCCUUUGCCACAAACUAAGCCCAUCC ....((((((....(((((......((((((((((((.......)))))))))))).-----)))))..))))))((((.......-......))))................ ( -37.62) >DroSec_CAF1 80934 106 + 1 GAACGAGGAGAUCGGAGCUACUUAACUGGCUAUGAGGAAAUUAGCCUCGUAGCCAGG-----AGCUGAACUUUUCUGGCCUU-UUC-CCCUUUGCAACAAACUAAGCCCAUCC ....((((((.((..((((......((((((((((((.......)))))))))))).-----)))))).))))))(((.(((-...-................))).)))... ( -32.91) >DroSim_CAF1 77458 107 + 1 GAACGAGUAGAUCGGAGCUACUUAACUGGCUAUGAGGAAAUUAGCCUCGUAGCCAGG-----AGCUGAGCUUUUCUGGCCUUUUUC-CCCUUUGCAACAAUCUAAGCCCAUCC (((.(((((((..((((((......((((((((((((.......)))))))))))).-----.....))))))))))..))).)))-.......................... ( -32.70) >DroEre_CAF1 81154 106 + 1 GAACGAGGAGAUCGGAGGUACUUAACUGGCUAUGAGGAAAUUAGCCUCGUAGCCAGG-----AGCUACGCUUUUCUGGCCUU-UCC-CCCUUUGCACCAAGCGAAGUCCAUCC ....(..(((..(((((........((((((((((((.......))))))))))))(-----(((...)))))))))..)))-..)-..((((((.....))))))....... ( -38.60) >DroYak_CAF1 82756 107 + 1 GAACGAGGAGAUCGCUGGUACUUAACUGGCUAUGAGGAAAUUAGCCUCGUGGCCAGG-----AGCUAAGCUUUUCUGGCCUU-UUCCCCUUUUGCACCCAGCUAAGACCAUCC ......(((((..((..(.......((((((((((((.......))))))))))))(-----(((...))))..)..))..)-)))).(((..((.....)).)))....... ( -35.80) >DroAna_CAF1 75667 102 + 1 GGAGGAGG----CGGAGAUAGAU-GCUGGCCAUGAGGAAAUUAGCCUCUUAGCCAAGAACAGAGCUUGGCUUUUCUGGCCUGUUAC-CGCUUC-----AUACUGAGCUAAUCC ....((((----(((.(((((..-((..(....((((.......))))..(((((((.......)))))))...)..))))))).)-))))))-----............... ( -34.80) >consensus GAACGAGGAGAUCGGAGCUACUUAACUGGCUAUGAGGAAAUUAGCCUCGUAGCCAGG_____AGCUAAGCUUUUCUGGCCUU_UUC_CCCUUUGCAACAAACUAAGCCCAUCC ...(((((.................((((((((((((.......)))))))))))).......((((........))))..........)))))................... (-22.52 = -23.22 + 0.70)


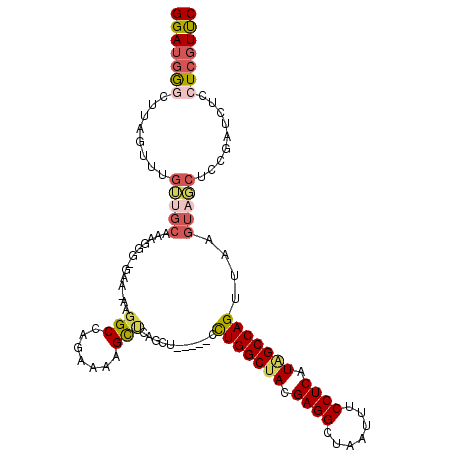
| Location | 10,948,589 – 10,948,696 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 82.90 |
| Mean single sequence MFE | -34.95 |
| Consensus MFE | -22.85 |
| Energy contribution | -23.75 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.65 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10948589 107 - 23771897 GGAUGGGCUUAGUUUGUGGCAAAGGG-GCAAAAGGCCAGAAAAGCUGAGCU-----CCUGGCUACGAGGCUAAUUUCCUCAUAGCCAGUUAAGUAGCUCCGAUCUCCUCGUUC (((.(((((((((((.((((......-.......))))...))))))))))-----)(((((((.((((.......)))).))))))).........)))............. ( -38.22) >DroSec_CAF1 80934 106 - 1 GGAUGGGCUUAGUUUGUUGCAAAGGG-GAA-AAGGCCAGAAAAGUUCAGCU-----CCUGGCUACGAGGCUAAUUUCCUCAUAGCCAGUUAAGUAGCUCCGAUCUCCUCGUUC ((((((((((..(((.((....)).)-)).-.))))).....((.((((((-----((((((((.((((.......)))).)))))))....).))))..)).))...))))) ( -28.90) >DroSim_CAF1 77458 107 - 1 GGAUGGGCUUAGAUUGUUGCAAAGGG-GAAAAAGGCCAGAAAAGCUCAGCU-----CCUGGCUACGAGGCUAAUUUCCUCAUAGCCAGUUAAGUAGCUCCGAUCUACUCGUUC (((((((..(((((((..((..((((-(.....(((.......)))...))-----)))(((((.((((.......)))).))))).........))..)))))))))))))) ( -36.40) >DroEre_CAF1 81154 106 - 1 GGAUGGACUUCGCUUGGUGCAAAGGG-GGA-AAGGCCAGAAAAGCGUAGCU-----CCUGGCUACGAGGCUAAUUUCCUCAUAGCCAGUUAAGUACCUCCGAUCUCCUCGUUC .((.(((..(((...(((((...(((-((.-.(.((.......)).)..))-----)))(((((.((((.......)))).)))))......)))))..)))..))))).... ( -37.60) >DroYak_CAF1 82756 107 - 1 GGAUGGUCUUAGCUGGGUGCAAAAGGGGAA-AAGGCCAGAAAAGCUUAGCU-----CCUGGCCACGAGGCUAAUUUCCUCAUAGCCAGUUAAGUACCAGCGAUCUCCUCGUUC ...(((((((((((((.((...........-..((((((...(((...)))-----.))))))..((((.......)))).)).))))))))).))))((((.....)))).. ( -38.20) >DroAna_CAF1 75667 102 - 1 GGAUUAGCUCAGUAU-----GAAGCG-GUAACAGGCCAGAAAAGCCAAGCUCUGUUCUUGGCUAAGAGGCUAAUUUCCUCAUGGCCAGC-AUCUAUCUCCG----CCUCCUCC (((...((..((...-----((.(((-....).(((((....(((((((.......)))))))..((((.......)))).))))).))-.))...))..)----).)))... ( -30.40) >consensus GGAUGGGCUUAGUUUGUUGCAAAGGG_GAA_AAGGCCAGAAAAGCUCAGCU_____CCUGGCUACGAGGCUAAUUUCCUCAUAGCCAGUUAAGUAGCUCCGAUCUCCUCGUUC (((((((........(((((.............(((.......)))...........(((((((.((((.......)))).)))))))....))))).........))))))) (-22.85 = -23.75 + 0.89)


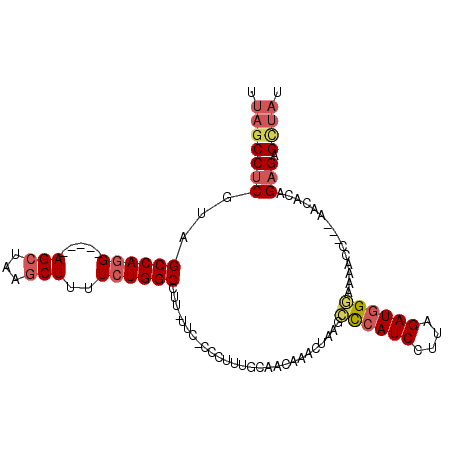
| Location | 10,948,629 – 10,948,726 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 78.89 |
| Mean single sequence MFE | -27.69 |
| Consensus MFE | -19.46 |
| Energy contribution | -20.27 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.956687 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10948629 97 + 23771897 UUAGCCUCGUAGCCAGG-----AGCUCAGCUUUUCUGGCCUUUUGC-CCCUUUGCCACAAACUAAGCCCAUCCUUAGAUGGGAAAACC---AACACAGAGAGCUAU .(((((((((.((.(((-----.((...(((.....))).....))-.)))..)).))........((((((....))))))......---......))).)))). ( -27.40) >DroSec_CAF1 80974 96 + 1 UUAGCCUCGUAGCCAGG-----AGCUGAACUUUUCUGGCCUU-UUC-CCCUUUGCAACAAACUAAGCCCAUCCUUAGAUGGGAAAACC---AACACAGAGAGCUAU .(((((((...((((((-----((.......)))))))).((-(((-((.((((...))))(((((......)))))..)))))))..---......))).)))). ( -27.60) >DroSim_CAF1 77498 97 + 1 UUAGCCUCGUAGCCAGG-----AGCUGAGCUUUUCUGGCCUUUUUC-CCCUUUGCAACAAUCUAAGCCCAUCCUUAGAUGGGAAAACC---AACACAGAGAGCUAU .(((((((...((((((-----(((...))))..)))))..(((((-((..........(((((((......))))))))))))))..---......))).)))). ( -29.80) >DroEre_CAF1 81194 96 + 1 UUAGCCUCGUAGCCAGG-----AGCUACGCUUUUCUGGCCUU-UCC-CCCUUUGCACCAAGCGAAGUCCAUCCUUAGAUGGUAAAACC---CACACAGAGAGUUAA .(((((((...((((((-----(((...))))..)))))...-...-..((((((.....)))))).(((((....))))).......---......))).)))). ( -26.40) >DroYak_CAF1 82796 97 + 1 UUAGCCUCGUGGCCAGG-----AGCUAAGCUUUUCUGGCCUU-UUCCCCUUUUGCACCCAGCUAAGACCAUCCUUAGAUGGUUUGCCC---GACACAGAGAGUUAA .(((((((..(((((((-----(((...))))..))))))..-..........(((.(((.(((((......))))).)))..)))..---......))).)))). ( -29.50) >DroAna_CAF1 75702 96 + 1 UUAGCCUCUUAGCCAAGAACAGAGCUUGGCUUUUCUGGCCUGUUAC-CGCUUC-----AUACUGAGCUAAUCCUUGGAUGGGCUUCCUCAAAACACUGCGAG---- ...(((....(((((((.......))))))).....))).......-(((...-----....((((..(..((......))..)..)))).......)))..---- ( -25.44) >consensus UUAGCCUCGUAGCCAGG_____AGCUAAGCUUUUCUGGCCUU_UUC_CCCUUUGCAACAAACUAAGCCCAUCCUUAGAUGGGAAAACC___AACACAGAGAGCUAU .(((((((...((((((.....(((...)))..))))))...........................((((((....))))))...............))).)))). (-19.46 = -20.27 + 0.81)


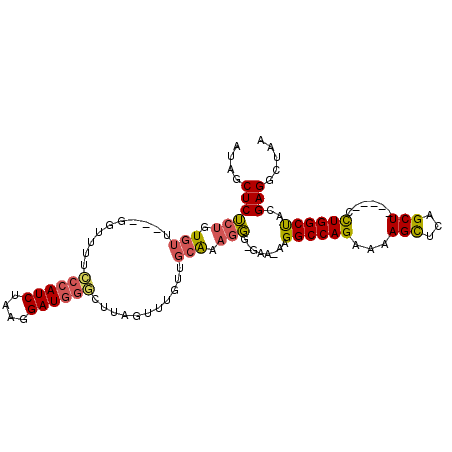
| Location | 10,948,629 – 10,948,726 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 78.89 |
| Mean single sequence MFE | -30.35 |
| Consensus MFE | -19.64 |
| Energy contribution | -19.75 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.744721 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10948629 97 - 23771897 AUAGCUCUCUGUGUU---GGUUUUCCCAUCUAAGGAUGGGCUUAGUUUGUGGCAAAGGG-GCAAAAGGCCAGAAAAGCUGAGCU-----CCUGGCUACGAGGCUAA .((((.(((......---......((((((....))))))........(((((..((((-((....(((.......)))..)))-----))).)))))))))))). ( -35.50) >DroSec_CAF1 80974 96 - 1 AUAGCUCUCUGUGUU---GGUUUUCCCAUCUAAGGAUGGGCUUAGUUUGUUGCAAAGGG-GAA-AAGGCCAGAAAAGUUCAGCU-----CCUGGCUACGAGGCUAA .((((.(((......---..(((((((..(((((......)))))((((...)))).))-)))-))((((((...((.....))-----.))))))..))))))). ( -28.20) >DroSim_CAF1 77498 97 - 1 AUAGCUCUCUGUGUU---GGUUUUCCCAUCUAAGGAUGGGCUUAGAUUGUUGCAAAGGG-GAAAAAGGCCAGAAAAGCUCAGCU-----CCUGGCUACGAGGCUAA .((((.(((......---..((((((((((((((......)))))))..........))-))))).((((((...(((...)))-----.))))))..))))))). ( -30.90) >DroEre_CAF1 81194 96 - 1 UUAACUCUCUGUGUG---GGUUUUACCAUCUAAGGAUGGACUUCGCUUGGUGCAAAGGG-GGA-AAGGCCAGAAAAGCGUAGCU-----CCUGGCUACGAGGCUAA ....((((((.(((.---.......(((((....)))))(((......)))))).))))-)).-..((((.......((((((.-----....)))))).)))).. ( -29.30) >DroYak_CAF1 82796 97 - 1 UUAACUCUCUGUGUC---GGGCAAACCAUCUAAGGAUGGUCUUAGCUGGGUGCAAAAGGGGAA-AAGGCCAGAAAAGCUUAGCU-----CCUGGCCACGAGGCUAA ....((((.((..((---.(((((((((((....)))))).)).))).))..))..))))...-..((((((...(((...)))-----.)))))).......... ( -33.40) >DroAna_CAF1 75702 96 - 1 ----CUCGCAGUGUUUUGAGGAAGCCCAUCCAAGGAUUAGCUCAGUAU-----GAAGCG-GUAACAGGCCAGAAAAGCCAAGCUCUGUUCUUGGCUAAGAGGCUAA ----.((((..(((.(((((.((..((......)).))..))))).))-----)..)))-).....((((.....(((((((.......)))))))....)))).. ( -24.80) >consensus AUAGCUCUCUGUGUU___GGUUUUCCCAUCUAAGGAUGGGCUUAGUUUGUUGCAAAGGG_GAA_AAGGCCAGAAAAGCUCAGCU_____CCUGGCUACGAGGCUAA ....((((((.(((..........((((((....))))))...........))).)))........((((((...(((...)))......))))))..)))..... (-19.64 = -19.75 + 0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:06:45 2006