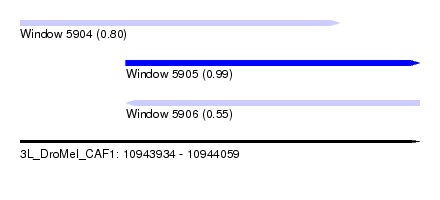
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,943,934 – 10,944,059 |
| Length | 125 |
| Max. P | 0.991853 |

| Location | 10,943,934 – 10,944,034 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 79.21 |
| Mean single sequence MFE | -27.85 |
| Consensus MFE | -17.91 |
| Energy contribution | -18.30 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.797015 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10943934 100 + 23771897 UUUCUCAUUCCGCUGAUUUGUGGUCGCGGCGAGGCGAAUGUGUGGCA-AAAAUGAUUUUAGCCGCAGAUUCAUAAAAUAUCUGCCCACAAUGGGUUGG--UGG ..((((...((((.(((.....))))))).)))).((((.(((((((-(((....)))).)))))).))))......((((.((((.....)))).))--)). ( -33.90) >DroVir_CAF1 68827 85 + 1 UAUCUCAUGCCGUUGAUUUGUGGUCGA--C----AAAAUAUGUGAUGCAAAAUUAUUUCAGCCGCAUAUUCAG-----AGCUGCCCACAACGGACA------- .......((((((((...((.(((.(.--.----..((((((((.((.(((....)))))..))))))))...-----..).))))))))))).))------- ( -18.70) >DroGri_CAF1 59028 85 + 1 UUUCUCAUGCCGUUGAUUUGUGGUCGA--C----AAAAUAUGUGAUGCAAAAUUAUUUCAGCCGCAUAUUCAG-----AGCUGCCCACAAUGGACA------- .......((((((((...((.(((.(.--.----..((((((((.((.(((....)))))..))))))))...-----..).))))))))))).))------- ( -16.90) >DroEre_CAF1 76693 94 + 1 UUUCUCAUUCCGCUGAUUUGUGGUCGCGGCGAGGCGAGUGUGUGGCA-AAAAUGAUUUUAGCCGCAUAUUCAUAAAAUAUCUGCCCACAAUGGGG-------- ..((((...((((.(((.....))))))).)))).((((((((((((-(((....)))).)))))))))))............(((.....))).-------- ( -31.50) >DroYak_CAF1 77748 94 + 1 UUUCUCAUUCCGCUGAUUUGUGGUCGCGGCGAGGCGAAUGUGUGGCA-AAAAUGAUUUUAGCCGCAUAUUCAUAAAAUAUCUGCCCACAAUGGGG-------- ..((((...((((.(((.....))))))).)))).((((((((((((-(((....)))).)))))))))))............(((.....))).-------- ( -31.50) >DroAna_CAF1 71310 102 + 1 UUUCUCAUUCCGCUGAUUUGUGGUCGAGGCGAGGCGAAUGUGUGGCA-AAAAUGAUUUCAGCCGCAGAUUCAUAAAAUAUCUGCCCACAAUGGGGUGGGCUGC ..((((...((((......))))..))))....((((((.((((((.-............)))))).))))...........((((((......)))))).)) ( -34.62) >consensus UUUCUCAUUCCGCUGAUUUGUGGUCGAGGCGAGGCGAAUGUGUGGCA_AAAAUGAUUUCAGCCGCAUAUUCAUAAAAUAUCUGCCCACAAUGGGG________ .........(((.....((((((.((.........(((((((((((..............)))))))))))..........)).))))))))).......... (-17.91 = -18.30 + 0.39)



| Location | 10,943,967 – 10,944,059 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 74.88 |
| Mean single sequence MFE | -29.62 |
| Consensus MFE | -16.20 |
| Energy contribution | -16.83 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.55 |
| SVM decision value | 2.29 |
| SVM RNA-class probability | 0.991853 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10943967 92 + 23771897 GCGAAUGUGUGGCA-AAAAUGAUUUUAGCCGCAGAUUCAUAAAAUAUCUGCCCACAAUGGGUUGGUGGUGGGCC-UGCUAGUGAUGC------UGCUGGC ..((((.(((((((-(((....)))).)))))).))))...........((((((.((......)).)))))).-.((((((.....------.)))))) ( -31.90) >DroVir_CAF1 68855 74 + 1 -AAAAUAUGUGAUGCAAAAUUAUUUCAGCCGCAUAUUCAG-----AGCUGCCCACAACGGACA------CCGCAGCACUAGAG---C------CG----- -..((((((((.((.(((....)))))..)))))))).((-----.(((((((.....))...------..))))).))....---.------..----- ( -14.50) >DroSec_CAF1 76297 92 + 1 GCGAAUGUGUGGCA-AAAAUGAUUUUAGCCGCAGAUUCAUAAAAUAUCUGUCCACAAUGGGGUGGUGGUGGGCU-GGCUAGUGGUGC------UGCUGGC ((((((.(((((((-(((....)))).)))))).))))......((((...((((......)))).)))).)).-.((((((.....------.)))))) ( -30.10) >DroSim_CAF1 72436 92 + 1 GCGAAUGUGUGGCA-AAAAUGAUUUUAGCCGCAGAUUCAUAAAAUAUCUGCCCACAAUGGGGUGGUGGUGGGCU-GGCUAGUGGUAC------UGCUGGC ..((((.(((((((-(((....)))).)))))).))))...........((((((.((......)).)))))).-.((((((.....------.)))))) ( -32.30) >DroEre_CAF1 76726 77 + 1 GCGAGUGUGUGGCA-AAAAUGAUUUUAGCCGCAUAUUCAUAAAAUAUCUGCCCACAAUGGGG------UGGGCU-UGC---------------UGCUGGC ((((((((((((((-(((....)))).)))))))))))...........((((((......)------))))).-.))---------------....... ( -30.40) >DroYak_CAF1 77781 92 + 1 GCGAAUGUGUGGCA-AAAAUGAUUUUAGCCGCAUAUUCAUAAAAUAUCUGCCCACAAUGGGG------UGGGCU-UGCUGGUGGUGCUGCUGCUGCUGGC ..((((((((((((-(((....)))).)))))))))))...........((((((......)------))))).-.((..(..((......))..)..)) ( -38.50) >consensus GCGAAUGUGUGGCA_AAAAUGAUUUUAGCCGCAGAUUCAUAAAAUAUCUGCCCACAAUGGGGU_____UGGGCU_UGCUAGUGGUGC______UGCUGGC ((((((.((((((..............)))))).))))...........(((((..............))))).....................)).... (-16.20 = -16.83 + 0.63)



| Location | 10,943,967 – 10,944,059 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 74.88 |
| Mean single sequence MFE | -20.47 |
| Consensus MFE | -11.24 |
| Energy contribution | -11.96 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.545768 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10943967 92 - 23771897 GCCAGCA------GCAUCACUAGCA-GGCCCACCACCAACCCAUUGUGGGCAGAUAUUUUAUGAAUCUGCGGCUAAAAUCAUUUU-UGCCACACAUUCGC ((.((..------......)).)).-.((((((............))))))...........((((.((.(((.((((....)))-)))).)).)))).. ( -21.80) >DroVir_CAF1 68855 74 - 1 -----CG------G---CUCUAGUGCUGCGG------UGUCCGUUGUGGGCAGCU-----CUGAAUAUGCGGCUGAAAUAAUUUUGCAUCACAUAUUUU- -----((------(---(......)))).((------(((..(((((...(((((-----(.......).)))))..)))))...))))).........- ( -17.10) >DroSec_CAF1 76297 92 - 1 GCCAGCA------GCACCACUAGCC-AGCCCACCACCACCCCAUUGUGGACAGAUAUUUUAUGAAUCUGCGGCUAAAAUCAUUUU-UGCCACACAUUCGC ((..(((------(.....)).)).-.))......((((......)))).............((((.((.(((.((((....)))-)))).)).)))).. ( -17.10) >DroSim_CAF1 72436 92 - 1 GCCAGCA------GUACCACUAGCC-AGCCCACCACCACCCCAUUGUGGGCAGAUAUUUUAUGAAUCUGCGGCUAAAAUCAUUUU-UGCCACACAUUCGC ....(((------(.....)).)).-.((((((............))))))...........((((.((.(((.((((....)))-)))).)).)))).. ( -21.50) >DroEre_CAF1 76726 77 - 1 GCCAGCA---------------GCA-AGCCCA------CCCCAUUGUGGGCAGAUAUUUUAUGAAUAUGCGGCUAAAAUCAUUUU-UGCCACACACUCGC ....((.---------------((.-.(((((------(......))))))..((((((...))))))))(((.((((....)))-))))........)) ( -20.30) >DroYak_CAF1 77781 92 - 1 GCCAGCAGCAGCAGCACCACCAGCA-AGCCCA------CCCCAUUGUGGGCAGAUAUUUUAUGAAUAUGCGGCUAAAAUCAUUUU-UGCCACACAUUCGC ((..((....)).)).......((.-.(((((------(......))))))...........((((.((.(((.((((....)))-)))).)).)))))) ( -25.00) >consensus GCCAGCA______GCACCACUAGCA_AGCCCA_____ACCCCAUUGUGGGCAGAUAUUUUAUGAAUAUGCGGCUAAAAUCAUUUU_UGCCACACAUUCGC ...........................(((((..............)))))...........((((.((.(((..............))).)).)))).. (-11.24 = -11.96 + 0.72)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:06:40 2006