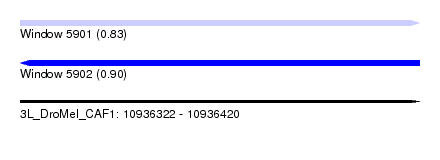
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,936,322 – 10,936,420 |
| Length | 98 |
| Max. P | 0.904329 |

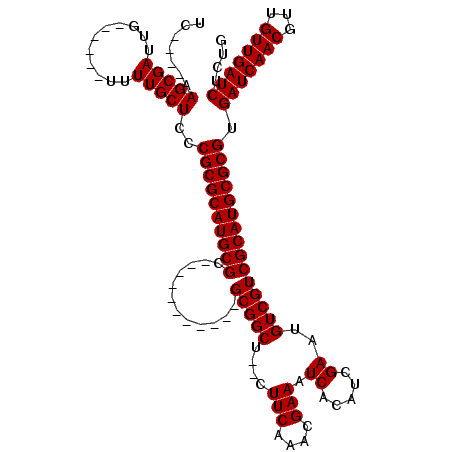
| Location | 10,936,322 – 10,936,420 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.21 |
| Mean single sequence MFE | -39.25 |
| Consensus MFE | -31.57 |
| Energy contribution | -31.57 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.826962 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10936322 98 + 23771897 UC-----AGCGAUUG------UUUUGCUCCCGCGCAUGCGC-----------GCGGCUCUCUUCAAACGAAAUCACAUCGAAUGUCGUCGCAUGCGCGUGAUCAACGUUGUUGAUCUCUA ..-----(((((...------..)))))..((((((((((.-----------(((((..((..................))..))))))))))))))).(((((((...))))))).... ( -36.07) >DroVir_CAF1 62668 97 + 1 UC----AAGCGAUUG------UUUUGCUCCCGCGCAUGCGC-----------GCGGCG--CUUCAAACGAAAUCACAUUGAAUGUCGUCGCAUGCGCGUGAUCAACGUUGUUGAUCUGUG ..----.(((((...------..)))))..((((((((((.-----------((((((--.(((((...........))))))))))))))))))))).(((((((...))))))).... ( -39.40) >DroPse_CAF1 72953 120 + 1 UCGUCGAAGCGAUUGUUCGGGUUUUGCUCCCGCGCAUGCGCCUUGCGGAGGAGCGGCUCUCUUCAAACGAAAUCACAUCGAAUGUCGUCGCAUGCGCGUGAUCAACGUUGUUGAUCUCUG ((((....))))......(((.......)))(((((((((.......((((((.....))))))..((((..((.....))...)))))))))))))(.(((((((...))))))).).. ( -40.40) >DroGri_CAF1 52881 97 + 1 UC----AAGCGAUUG------UUUUGCUCCCGCGCAUGCGC-----------GCGGCA--UUUCAAACGAAAUCACAUUGAAUGUCGUCGCAUGCGCGUGAUCAACGUUGUUGAUCUGUG ..----.(((((...------..)))))..((((((((((.-----------((((((--((.(((...........))))))))))))))))))))).(((((((...))))))).... ( -39.80) >DroMoj_CAF1 67470 97 + 1 UC----AAGCGAUUG------UUUUGCUCCCGCGCAUGCGC-----------GCGGCG--CUUCAAACGAAAUCACAUUGAAUGUCGUCGCAUGCGCGUGAUCAACGUUGUUGAUCUGUG ..----.(((((...------..)))))..((((((((((.-----------((((((--.(((((...........))))))))))))))))))))).(((((((...))))))).... ( -39.40) >DroPer_CAF1 60458 120 + 1 UCGUCGAAGCGAUUGUUCGGGUUUUGCUCCCGCGCAUGCGCCUUGCGGAGGAGCGGCUCUCUUCAAACGAAAUCACAUCGAAUGUCGUCGCAUGCGCGUGAUCAACGUUGUUGAUCUCUG ((((....))))......(((.......)))(((((((((.......((((((.....))))))..((((..((.....))...)))))))))))))(.(((((((...))))))).).. ( -40.40) >consensus UC____AAGCGAUUG______UUUUGCUCCCGCGCAUGCGC___________GCGGCU__CUUCAAACGAAAUCACAUCGAAUGUCGUCGCAUGCGCGUGAUCAACGUUGUUGAUCUCUG .......(((((...........)))))..((((((((((............(((((....(((....))).((.....))..))))))))))))))).(((((((...))))))).... (-31.57 = -31.57 + -0.00)

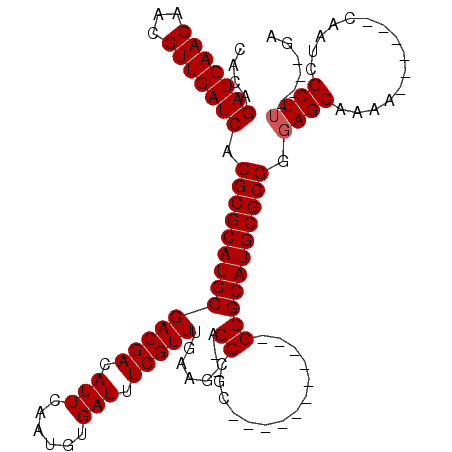
| Location | 10,936,322 – 10,936,420 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.21 |
| Mean single sequence MFE | -37.30 |
| Consensus MFE | -31.15 |
| Energy contribution | -31.32 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.904329 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10936322 98 - 23771897 UAGAGAUCAACAACGUUGAUCACGCGCAUGCGACGACAUUCGAUGUGAUUUCGUUUGAAGAGAGCCGC-----------GCGCAUGCGCGGGAGCAAAA------CAAUCGCU-----GA ....(((((((...))))))).((((((((((.((.....))..(((.((((.......))))..)))-----------.))))))))))..(((....------.....)))-----.. ( -35.00) >DroVir_CAF1 62668 97 - 1 CACAGAUCAACAACGUUGAUCACGCGCAUGCGACGACAUUCAAUGUGAUUUCGUUUGAAG--CGCCGC-----------GCGCAUGCGCGGGAGCAAAA------CAAUCGCUU----GA ....(((((((...))))))).((((((((((.((...(((((((......)).))))).--.....)-----------))))))))))).((((....------.....))))----.. ( -35.30) >DroPse_CAF1 72953 120 - 1 CAGAGAUCAACAACGUUGAUCACGCGCAUGCGACGACAUUCGAUGUGAUUUCGUUUGAAGAGAGCCGCUCCUCCGCAAGGCGCAUGCGCGGGAGCAAAACCCGAACAAUCGCUUCGACGA ....(((((((...))))))).((((((((((.........((.(((.((((.......))))..)))))..((....))))))))))))(((((...............)))))..... ( -41.36) >DroGri_CAF1 52881 97 - 1 CACAGAUCAACAACGUUGAUCACGCGCAUGCGACGACAUUCAAUGUGAUUUCGUUUGAAA--UGCCGC-----------GCGCAUGCGCGGGAGCAAAA------CAAUCGCUU----GA ....(((((((...))))))).((((((((((.((.(((((((((......)).))).))--))...)-----------))))))))))).((((....------.....))))----.. ( -35.50) >DroMoj_CAF1 67470 97 - 1 CACAGAUCAACAACGUUGAUCACGCGCAUGCGACGACAUUCAAUGUGAUUUCGUUUGAAG--CGCCGC-----------GCGCAUGCGCGGGAGCAAAA------CAAUCGCUU----GA ....(((((((...))))))).((((((((((.((...(((((((......)).))))).--.....)-----------))))))))))).((((....------.....))))----.. ( -35.30) >DroPer_CAF1 60458 120 - 1 CAGAGAUCAACAACGUUGAUCACGCGCAUGCGACGACAUUCGAUGUGAUUUCGUUUGAAGAGAGCCGCUCCUCCGCAAGGCGCAUGCGCGGGAGCAAAACCCGAACAAUCGCUUCGACGA ....(((((((...))))))).((((((((((.........((.(((.((((.......))))..)))))..((....))))))))))))(((((...............)))))..... ( -41.36) >consensus CACAGAUCAACAACGUUGAUCACGCGCAUGCGACGACAUUCAAUGUGAUUUCGUUUGAAG__AGCCGC___________GCGCAUGCGCGGGAGCAAAA______CAAUCGCUU____GA ....(((((((...))))))).((((((((((((((.(((......))).)))))........((..............))))))))))).((((...............))))...... (-31.15 = -31.32 + 0.17)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:06:36 2006