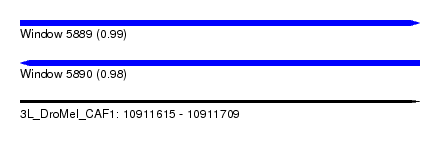
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,911,615 – 10,911,709 |
| Length | 94 |
| Max. P | 0.986458 |

| Location | 10,911,615 – 10,911,709 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.94 |
| Mean single sequence MFE | -29.77 |
| Consensus MFE | -19.87 |
| Energy contribution | -20.37 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.67 |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.986458 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
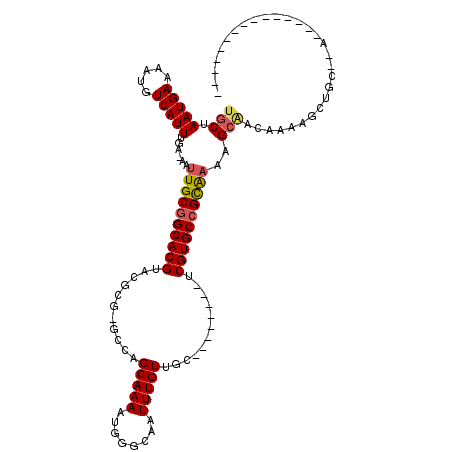
>3L_DroMel_CAF1 10911615 94 + 23771897 ---------------U--UCAUCCUUUGCCGCUUUUACGGCACGA--------GCAACAAAUUGCCCAUUUUGCAGGC-CUCGUACGUGCGGCAAUUUUCGAAUGACAUUUUCAUUAGCA ---------------.--.......(((((((....(((..((((--------((..((((........))))...).-))))).))))))))))......(((((.....))))).... ( -23.50) >DroVir_CAF1 34428 92 + 1 ------------------GCUGCUGCCGUUGCACACGCGGCACGA--------GCAACAAAUUGCCCAUUUUGCUGGC-CGCGUACGUGCCGCAAUU-UCAAAUGACAUUUUCAUUAGCA ------------------...(((...((.(((((((((((.(.(--------((((.............))))))))-)))))..)))).))....-...(((((.....)))))))). ( -29.22) >DroPse_CAF1 47031 97 + 1 -----------------------UUUUAUUCCUUUUGCGGCACGAGGCAGCCAGCAACAAAUUGCCCAUUUUGCAGCCUCUGGUACGUGCCGCAAUUUUCAAAUGACAUUUUCAUUAGCA -----------------------...........((((((((((.(.(((...((..((((........))))..))..))).).))))))))))......(((((.....))))).... ( -27.60) >DroGri_CAF1 31499 108 + 1 GAACUAUGUGGCUGGU--GUUGCUGUUGCUGCACUCGCGGCACGA--------GCAACAAAUUGCCCAUUUUGCUGGU-CGCGUACGUGCCGCAAUU-UCAAAUGACAUUUUCAUUAGCA (((..(((.(((...(--((((((..((((((....))))))..)--------))))))....)))))).((((.((.-(((....))))))))).)-)).(((((.....))))).... ( -37.00) >DroMoj_CAF1 42618 96 + 1 -AAC-----------C--GCAGCUGCUGUUGCACUCGCGGCACGA--------GCAACAAAUUGCCCAUUUUGCUGGC-CGCGUACGUGCCGCAAUU-UCAAAUGACAUUUUCAUUAGCA -...-----------.--(((((....)))))....((((((((.--------((((....))))(((......))).-......))))))))....-...(((((.....))))).... ( -29.70) >DroAna_CAF1 35618 96 + 1 ---------------UUGUCAUCUUUGGCGGCUUUUGCGGCACGA--------GCAACAAAUUGCCCAUUUUGCAAAC-CGUGUACGUGCCGCAAUUUUCAAAUGACAUUUUCAUUAGCA ---------------.(((((....)))))(((.((((((((((.--------(((.....((((.......))))..-..))).))))))))))......(((((.....)))))))). ( -31.60) >consensus _______________U__GCAGCUGUUGCUGCACUCGCGGCACGA________GCAACAAAUUGCCCAUUUUGCAGGC_CGCGUACGUGCCGCAAUU_UCAAAUGACAUUUUCAUUAGCA ..............................((....((((((((.........((((....))))......(((........)))))))))))........(((((.....))))).)). (-19.87 = -20.37 + 0.50)



| Location | 10,911,615 – 10,911,709 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.94 |
| Mean single sequence MFE | -31.97 |
| Consensus MFE | -20.33 |
| Energy contribution | -20.50 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.984756 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10911615 94 - 23771897 UGCUAAUGAAAAUGUCAUUCGAAAAUUGCCGCACGUACGAG-GCCUGCAAAAUGGGCAAUUUGUUGC--------UCGUGCCGUAAAAGCGGCAAAGGAUGA--A--------------- ..............((((((.....(((((((...(((..(-((.......((((((((....))))--------))))))))))...))))))).))))))--.--------------- ( -29.31) >DroVir_CAF1 34428 92 - 1 UGCUAAUGAAAAUGUCAUUUGA-AAUUGCGGCACGUACGCG-GCCAGCAAAAUGGGCAAUUUGUUGC--------UCGUGCCGCGUGUGCAACGGCAGCAGC------------------ (((((((((.....)))))...-..(((..((((..(((((-((.......((((((((....))))--------)))))))))))))))..))).))))..------------------ ( -31.91) >DroPse_CAF1 47031 97 - 1 UGCUAAUGAAAAUGUCAUUUGAAAAUUGCGGCACGUACCAGAGGCUGCAAAAUGGGCAAUUUGUUGCUGGCUGCCUCGUGCCGCAAAAGGAAUAAAA----------------------- ..(.(((((.....))))).)....((((((((((...(((.(((.(((((........))))).)))..)))...))))))))))...........----------------------- ( -31.40) >DroGri_CAF1 31499 108 - 1 UGCUAAUGAAAAUGUCAUUUGA-AAUUGCGGCACGUACGCG-ACCAGCAAAAUGGGCAAUUUGUUGC--------UCGUGCCGCGAGUGCAGCAACAGCAAC--ACCAGCCACAUAGUUC (((((((((.....)))))((.-..(((((((((....((.-....)).....((((((....))))--------)))))))))))...)).....))))..--................ ( -32.50) >DroMoj_CAF1 42618 96 - 1 UGCUAAUGAAAAUGUCAUUUGA-AAUUGCGGCACGUACGCG-GCCAGCAAAAUGGGCAAUUUGUUGC--------UCGUGCCGCGAGUGCAACAGCAGCUGC--G-----------GUU- ..(.(((((.....))))).).-(((((((((.....((((-((.......((((((((....))))--------))))))))))..(((....))))))))--)-----------)))- ( -34.11) >DroAna_CAF1 35618 96 - 1 UGCUAAUGAAAAUGUCAUUUGAAAAUUGCGGCACGUACACG-GUUUGCAAAAUGGGCAAUUUGUUGC--------UCGUGCCGCAAAAGCCGCCAAAGAUGACAA--------------- ............((((((((.......(((((.((....))-.(((((...((((((((....))))--------))))...))))).)))))...)))))))).--------------- ( -32.60) >consensus UGCUAAUGAAAAUGUCAUUUGA_AAUUGCGGCACGUACGCG_GCCAGCAAAAUGGGCAAUUUGUUGC________UCGUGCCGCAAAAGCAACAAAAGCUGC__A_______________ (((.(((((.....)))))......((((((((((...........(((((........)))))............))))))))))..)))............................. (-20.33 = -20.50 + 0.17)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:06:23 2006