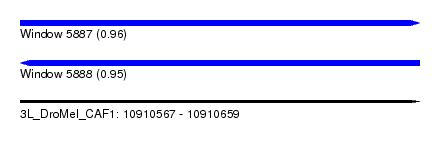
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,910,567 – 10,910,659 |
| Length | 92 |
| Max. P | 0.964047 |

| Location | 10,910,567 – 10,910,659 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 80.08 |
| Mean single sequence MFE | -31.47 |
| Consensus MFE | -21.47 |
| Energy contribution | -21.42 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.98 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.964047 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
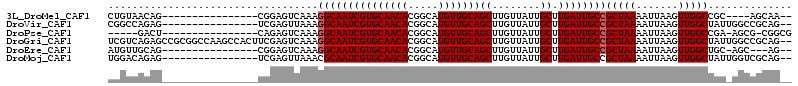
>3L_DroMel_CAF1 10910567 92 + 23771897 --UUGCU----GCGGCCAACUUAAUUUUAGCGGCAAUCAAGCAAUAACAAGCUGCAACAUGCCGUGUUGCACGAUUGCCUUUGACUCCG----------------CUGUUACAG --.((..----(((((...((.......)).(((((((.(((........)))(((((((...)))))))..))))))).........)----------------))))..)). ( -27.00) >DroVir_CAF1 33696 96 + 1 --CUGCGGCCAAUAGCCAACUUAAUUUUAGCGGCAAUCAAGCAAUAACAAGCUGCAACAUGCCGUGUUGCACGAUUGCCUUUAACUCGA----------------CUCUGGCCG --...((((((...((.............))(((((((.(((........)))(((((((...)))))))..)))))))..........----------------...)))))) ( -31.62) >DroPse_CAF1 45843 91 + 1 CGCCG-CGCU-UCGGCCAACUUAAUUUUAGCGGCAAUCAAGCAAUAACAAGCUGCAACAUGCCGUGUUGCACGAUUGCCUUUGACUCUG----------------AGUC----- .((((-....-.))))..(((((...((((.(((((((.(((........)))(((((((...)))))))..))))))).))))...))----------------))).----- ( -30.10) >DroGri_CAF1 30741 112 + 1 --CUGCGGCCAAUAGCCAACUUAAUUUUAGCGGCAAUCAAGCAAUAACAAGCUGCAACAUGCCGUGUUGCACGAUUGCCUUUGACUCGAAGUGGCUUGGCCGCGGCUCUGACGA --(((((((((..(((((.(((....((((.(((((((.(((........)))(((((((...)))))))..))))))).))))....)))))))))))))))))......... ( -49.60) >DroEre_CAF1 41953 92 + 1 --CU---GCU-GCAGCCAACUUAAUUUUAGCGGCAAUCAAGCAAUAACAAGCUGCAACAUGCCGUGUUGCACGAUUGCCUUUGACUCCG----------------CUGCAACAU --.(---(.(-(((((...((.......)).(((((((.(((........)))(((((((...)))))))..))))))).........)----------------))))).)). ( -30.30) >DroMoj_CAF1 41823 96 + 1 --CUGCGACCAAUAGCCAACUUAAUUUUAGCGGCAAUCAAGCAAUAACAAGCUGCAACAUGCCGUGUUGCACGAUUGCGUUUAACUCGA----------------CUCUGUCCA --..(.(((.......................((((((.(((........)))(((((((...)))))))..))))))(((......))----------------)...)))). ( -20.20) >consensus __CUGCGGCC_ACAGCCAACUUAAUUUUAGCGGCAAUCAAGCAAUAACAAGCUGCAACAUGCCGUGUUGCACGAUUGCCUUUGACUCGA________________CUCUGACAA ..........................((((.(((((((.(((........)))(((((((...)))))))..))))))).)))).............................. (-21.47 = -21.42 + -0.05)

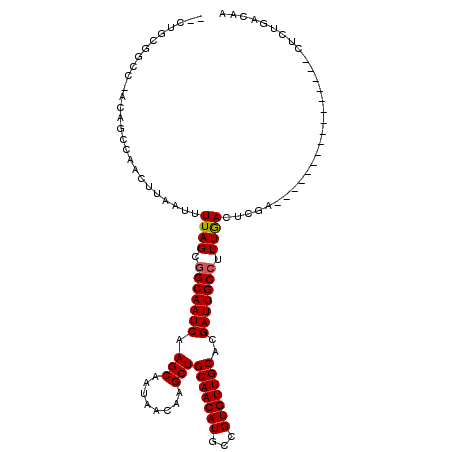
| Location | 10,910,567 – 10,910,659 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 80.08 |
| Mean single sequence MFE | -34.49 |
| Consensus MFE | -23.25 |
| Energy contribution | -23.42 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.949372 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10910567 92 - 23771897 CUGUAACAG----------------CGGAGUCAAAGGCAAUCGUGCAACACGGCAUGUUGCAGCUUGUUAUUGCUUGAUUGCCGCUAAAAUUAAGUUGGCCGC----AGCAA-- ........(----------------((........(((((((((((((((.....)))))))((........)).))))))))(((((.......))))))))----.....-- ( -28.80) >DroVir_CAF1 33696 96 - 1 CGGCCAGAG----------------UCGAGUUAAAGGCAAUCGUGCAACACGGCAUGUUGCAGCUUGUUAUUGCUUGAUUGCCGCUAAAAUUAAGUUGGCUAUUGGCCGCAG-- (((((((((----------------((((.((((.(((((((((((((((.....)))))))((........)).)))))))).......)))).)))))).)))))))...-- ( -39.70) >DroPse_CAF1 45843 91 - 1 -----GACU----------------CAGAGUCAAAGGCAAUCGUGCAACACGGCAUGUUGCAGCUUGUUAUUGCUUGAUUGCCGCUAAAAUUAAGUUGGCCGA-AGCG-CGGCG -----(((.----------------....)))...(((((((((((((((.....)))))))((........)).))))))))...............((((.-....-)))). ( -30.10) >DroGri_CAF1 30741 112 - 1 UCGUCAGAGCCGCGGCCAAGCCACUUCGAGUCAAAGGCAAUCGUGCAACACGGCAUGUUGCAGCUUGUUAUUGCUUGAUUGCCGCUAAAAUUAAGUUGGCUAUUGGCCGCAG-- ...........(((((((((((((((.........(((((((((((((((.....)))))))((........)).)))))))).........))).)))))..)))))))..-- ( -44.97) >DroEre_CAF1 41953 92 - 1 AUGUUGCAG----------------CGGAGUCAAAGGCAAUCGUGCAACACGGCAUGUUGCAGCUUGUUAUUGCUUGAUUGCCGCUAAAAUUAAGUUGGCUGC-AGC---AG-- .((((((((----------------(.........(((((((((((((((.....)))))))((........)).))))))))(((.......)))..)))))-)))---).-- ( -36.30) >DroMoj_CAF1 41823 96 - 1 UGGACAGAG----------------UCGAGUUAAACGCAAUCGUGCAACACGGCAUGUUGCAGCUUGUUAUUGCUUGAUUGCCGCUAAAAUUAAGUUGGCUAUUGGUCGCAG-- (((((..((----------------((((.((((..((((((((((((((.....)))))))((........)).)))))))........)))).))))))....))).)).-- ( -27.10) >consensus CGGUCAGAG________________CCGAGUCAAAGGCAAUCGUGCAACACGGCAUGUUGCAGCUUGUUAUUGCUUGAUUGCCGCUAAAAUUAAGUUGGCUAU_GGCCGCAG__ ...................................(((((((((((((((.....)))))))((........)).))))))))(((((.......))))).............. (-23.25 = -23.42 + 0.17)
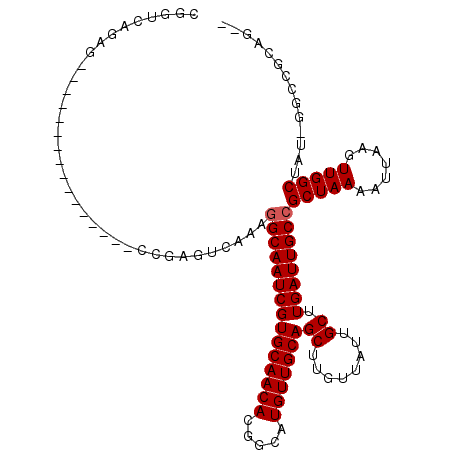
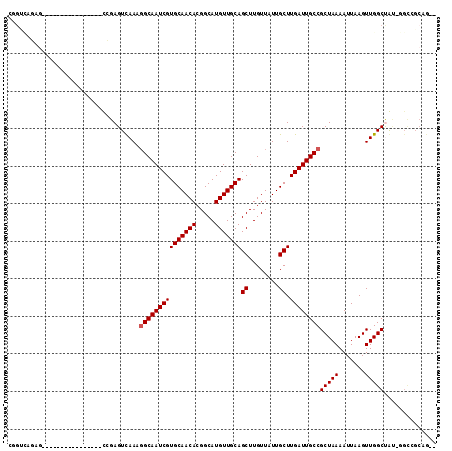
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:06:21 2006