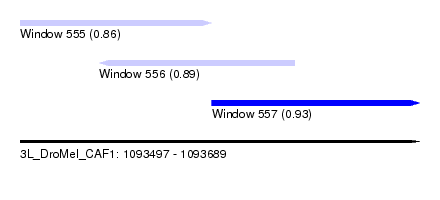
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,093,497 – 1,093,689 |
| Length | 192 |
| Max. P | 0.928488 |

| Location | 1,093,497 – 1,093,589 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.85 |
| Mean single sequence MFE | -27.94 |
| Consensus MFE | -18.16 |
| Energy contribution | -17.16 |
| Covariance contribution | -1.00 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.862218 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1093497 92 + 23771897 UGGCAAUGGCUUUGUGGACACACCCCAGAUAUCCCCGA----------------------------UGUUCCCCGAUAUAUACAUAUAGAGUAUGUUGGGUCCCCUCGAAGCCCGCUCCU .(((...(((((((.((..........((((((...))----------------------------)))).((((((((((.(.....).))))))))))...)).))))))).)))... ( -28.00) >DroSec_CAF1 10894 91 + 1 UGCCAAUGGCUUUGUGGACACACCCCAGAUAUC-CCGA----------------------------UGUCCCCCGAUAUAUACAUAUAGAGUAUGUUGGGUCCCCUCGAAGCCCGCUCCU .((....(((((((.((..........(((((.-...)----------------------------)))).((((((((((.(.....).))))))))))...)).))))))).)).... ( -27.80) >DroSim_CAF1 2826 92 + 1 UGCCAAUGGCUUUGUGGACACACCCCAGAUAUCCCCGA----------------------------UGUCCCCCGAUAUAUACAUAUAGAGUAUGUUGGGUCCCCUCGAAGCCCGCUCCU .((....(((((((.((..........((((((...))----------------------------)))).((((((((((.(.....).))))))))))...)).))))))).)).... ( -28.90) >DroEre_CAF1 4872 108 + 1 UGGCAAUGGCUUUGUGGACACACCCCAGAUAUCCCCGAUGUC---------CCCAAUGUCCCCCGAUGUCCCCCGAUAUAUGUAUAU--AGUGUGCUGUG-CCCCUCGAAGCCCGCUCCU .(((...(((((((.((.((((.....((((((...(((((.---------....)))))....))))))...........((((..--...))))))))-..)).))))))).)))... ( -25.90) >DroYak_CAF1 6549 118 + 1 UGGCAAUGGCUUUGUGGACACACCCCAGAUAUCCCCGAUAUCUCCGAUGCGCCCAAUAUCCCCCUAUAUCCCCCGAUAUAUGUAUAU--AGUAUGCUAUGUCCCCUCGAAGCCCGCUCCU .(((...(((((((.(((((......(((((((...))))))).....(((..(.((((.(...((((((....)))))).).))))--.)..)))..)))))...))))))).)))... ( -29.10) >consensus UGGCAAUGGCUUUGUGGACACACCCCAGAUAUCCCCGA____________________________UGUCCCCCGAUAUAUACAUAUAGAGUAUGUUGGGUCCCCUCGAAGCCCGCUCCU .......(((((((.((((...............................................((((....))))...((((((...))))))...))))...)))))))....... (-18.16 = -17.16 + -1.00)

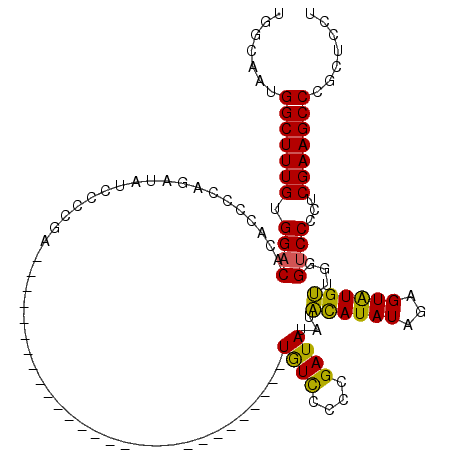
| Location | 1,093,535 – 1,093,629 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.52 |
| Mean single sequence MFE | -33.34 |
| Consensus MFE | -29.62 |
| Energy contribution | -29.78 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.886778 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1093535 94 - 23771897 UCCCUGCAGAUAUCUCCUCUUGCAGCCACUUGUCCUCCAAAGGAGCGGGCUUCGAGGGGACCCAACAUACUCUAUAUGUAUAUAUCGGGGAACA-------------------------- ((((((...((((.((((((((.((((.....((((....))))...)))).))))))))....(((((.....)))))))))..))))))...-------------------------- ( -32.70) >DroSec_CAF1 10931 94 - 1 UCCCUGCAGAUAUCUCCUCUUGCAGCCACUUGUCCUCCAAAGGAGCGGGCUUCGAGGGGACCCAACAUACUCUAUAUGUAUAUAUCGGGGGACA-------------------------- (((((...(((((.((((((((.((((.....((((....))))...)))).))))))))....(((((.....)))))..))))).)))))..-------------------------- ( -33.00) >DroSim_CAF1 2864 94 - 1 UCCCUGCAGAUAUCUCCUCUUGCAGCCACUUGUCCUCCAAAGGAGCGGGCUUCGAGGGGACCCAACAUACUCUAUAUGUAUAUAUCGGGGGACA-------------------------- (((((...(((((.((((((((.((((.....((((....))))...)))).))))))))....(((((.....)))))..))))).)))))..-------------------------- ( -33.00) >DroEre_CAF1 4912 108 - 1 UCCCUGCAGAUAUCUCCUCUUGCAGCCACUUGUCCUCAAAAGGAGCGGGCUUCGAGGGG-CACAGCACACU--AUAUACAUAUAUCGGGGGACAUCGGGGGACAUUGGG---------GA (((((...(((.((((((((((.((((.....((((....))))...)))).))))..(-(...)).....--.............)))))).))).(....)...)))---------)) ( -31.10) >DroYak_CAF1 6589 118 - 1 UCCCUGCAGAUAUCUCCUCUUGCAGCCACUUGUCCUCAAAAGGAGCGGGCUUCGAGGGGACAUAGCAUACU--AUAUACAUAUAUCGGGGGAUAUAGGGGGAUAUUGGGCGCAUCGGAGA (((.(((.((((((((((......((....(((((((....((((....))))..)))))))..)).....--.......((((((....))))))))))))))))....)))..))).. ( -36.90) >consensus UCCCUGCAGAUAUCUCCUCUUGCAGCCACUUGUCCUCCAAAGGAGCGGGCUUCGAGGGGACCCAACAUACUCUAUAUGUAUAUAUCGGGGGACA__________________________ ((((((...((((.((((((((.((((.....((((....))))...)))).))))))))....(((((.....)))))))))..))))))............................. (-29.62 = -29.78 + 0.16)

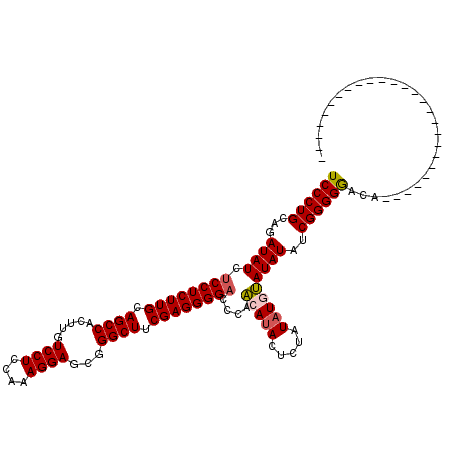
| Location | 1,093,589 – 1,093,689 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 83.07 |
| Mean single sequence MFE | -28.34 |
| Consensus MFE | -19.58 |
| Energy contribution | -20.38 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.928488 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1093589 100 + 23771897 UUGGAGGACAAGUGGCUGCAAGAGGAGAUAUCUGCAGGGAGACAAACACAGAACACAAACAGACGCCCACACACAUGCACACACGCAGG-G-----AUGUCCUUAU ...(((((((.(((((((((.((.......)))))))(....).......................)))).(...(((......)))..-)-----.))))))).. ( -26.60) >DroSec_CAF1 10985 99 + 1 UUGGAGGACAAGUGGCUGCAAGAGGAGAUAUCUGCAGGGAGACAAACACAGAACACAAAUAGACGCC-UCACACAGGCACACAUGCAGC-G-----AUGUCCUUAU ...(((((((..(.((((((..........((((...(....).....))))............(((-(.....)))).....))))))-.-----)))))))).. ( -30.40) >DroSim_CAF1 2918 99 + 1 UUGGAGGACAAGUGGCUGCAAGAGGAGAUAUCUGCAGGGAGACAAACACAGAACACAAACAGACGCC-UCACACAGGCACACAUGCAGC-G-----AUGUCCUUAU ...(((((((..(.((((((..........((((...(....).....))))............(((-(.....)))).....))))))-.-----)))))))).. ( -30.40) >DroEre_CAF1 4980 88 + 1 UUUGAGGACAAGUGGCUGCAAGAGGAGAUAUCUGCAGGGAGACAAACACAGAGCACAAACAGG------------------GAUGCAGGGAUGUGUGUGUCCUUAU ..((((((((.(((.(((((.((.......)))))))(....)...)))...(((((..(..(------------------....)..)..))))).)))))))). ( -28.40) >DroYak_CAF1 6667 93 + 1 UUUGAGGACAAGUGGCUGCAAGAGGAGAUAUCUGCAGGGAGACAAACACGGAGCACAAACAGACGCCCACA--------CACAUGCAAGGG-----AUGUCCUUAU ..((((((((.(((.(((((.((.......)))))))(....)...)))((.((..........))))...--------............-----.)))))))). ( -25.90) >consensus UUGGAGGACAAGUGGCUGCAAGAGGAGAUAUCUGCAGGGAGACAAACACAGAACACAAACAGACGCC_ACACACA_GCACACAUGCAGG_G_____AUGUCCUUAU ...(((((((.(((.(((((.((.......)))))))(....)...)))...........................((......))...........))))))).. (-19.58 = -20.38 + 0.80)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:40:28 2006