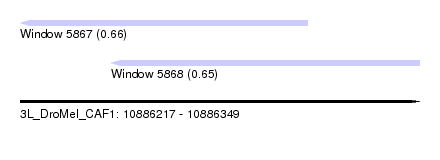
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,886,217 – 10,886,349 |
| Length | 132 |
| Max. P | 0.658874 |

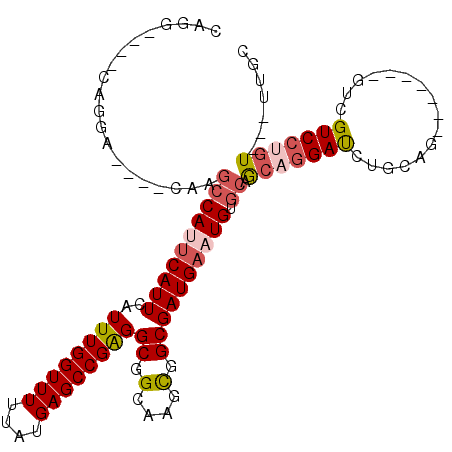
| Location | 10,886,217 – 10,886,312 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 80.54 |
| Mean single sequence MFE | -36.65 |
| Consensus MFE | -21.84 |
| Energy contribution | -22.71 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.658874 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10886217 95 - 23771897 CCAG----CAGGA----CGAGCCAUUCAUUCAUUUGGUUUUUAUGAGCCGAGGCGGCAAGCGGCGAUGAAUGUGCAGCAGGAUCUGCUG-------GUCGUCCUGU---UUGC ..((----(((((----(((((((((((((..((((((((....))))))))((.(....).)))))))))).))(((((...))))).-------.)))))))))---)... ( -41.50) >DroSec_CAF1 18554 95 - 1 CAGG----CAGGA----CUAGCCAUUCAUUCAUUUGGUUUUUAUGAGCCGAGGCGGCAAGCGGCGAUGAAUGUGCAGCAGGAUCUGCAG-------GUCGUCCUGU---UUGC ((((----(((((----(..((((((((((..((((((((....))))))))((.(....).)))))))))).)).((((...))))..-------...)))))))---))). ( -40.10) >DroSim_CAF1 14447 95 - 1 CAGG----CAGGA----CUAGCCAUUCAUUCAUUUGGUUUUUAUGAGCCGAGGCGGCAAGCGGCGAUGAAUGUGCAGCAGGAUCUGCAG-------GUCGUCCUGU---UUGC ((((----(((((----(..((((((((((..((((((((....))))))))((.(....).)))))))))).)).((((...))))..-------...)))))))---))). ( -40.10) >DroEre_CAF1 17469 102 - 1 CAGC----CAGCAGAAGCCAGCCACUCAUUCAUUUGGUUUUUAUGAGCCGAGGCGGCAAGCGGCGAUGAAUGUGCAGCAGGAUCUGCAG-------GUCGUCCAGUGUCCUGC ..((----(.(((((..((.((..(.((((((((((((((....))))))).((.(....).)).))))))).)..)).)).))))).)-------))....(((....))). ( -31.90) >DroYak_CAF1 15421 102 - 1 CAGCAGGCCAGGAGUAG-AAGCCAUUCAUUCAUUUGGUUUUUAUGAGCCGAGGCGGCGAGUGGCGAUGAAUGUGCAGCAGGAUCUGCAG-------GUCGUCCUGU---UUGC ......(((.((.((((-((((((..........))))))))))...))..))).(((((..(.(((((.(.(((((......))))).-------)))))))..)---)))) ( -37.80) >DroAna_CAF1 7720 103 - 1 CUGG----CAUAGAGAG-AGAGCAGCCAUUCAUUUGGUUUUUAUGAGCCGGGGCAGCAAGUGGCGAUGAAUGUGCUA--CGACCUGGACGAGGAUAGAGGUCCUGU---UUCU ....----...(((((.-((.(.(((((((((((((((((....)))))).....((.....)))))))))).))).--)(((((............))))))).)---)))) ( -28.50) >consensus CAGG____CAGGA____CAAGCCAUUCAUUCAUUUGGUUUUUAUGAGCCGAGGCGGCAAGCGGCGAUGAAUGUGCAGCAGGAUCUGCAG_______GUCGUCCUGU___UUGC ....................((((((((((..((((((((....))))))))((.(....).)))))))))).)).(((((((................)))))))....... (-21.84 = -22.71 + 0.86)


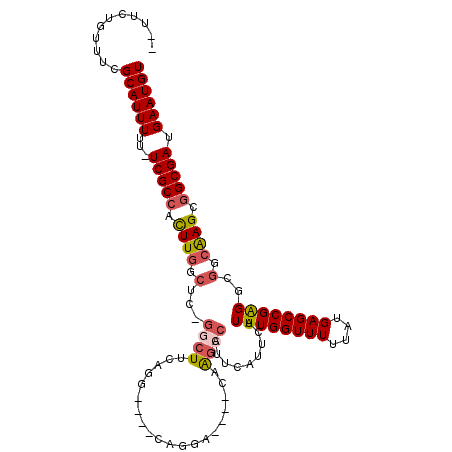
| Location | 10,886,247 – 10,886,349 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 84.65 |
| Mean single sequence MFE | -39.23 |
| Consensus MFE | -24.47 |
| Energy contribution | -25.08 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.30 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.654051 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10886247 102 - 23771897 --UUCUGUUUCGCAUUUUU-UCGCCACUUGGCUCAGGCUUCCAG----CAGGA----CGAGCCAUUCAUUCAUUUGGUUUUUAUGAGCCGAGGCGGCAAGCGGCGAUGAAUGU --.........((((((..-(((((.((((((((((((((((..----..)))----..))))..(((......)))......)))))))))((.....))))))).)))))) ( -39.60) >DroSec_CAF1 18584 102 - 1 --UUCUGUUUCGCAUUUUU-UCGCCACUUGGCUCAGGCUUCAGG----CAGGA----CUAGCCAUUCAUUCAUUUGGUUUUUAUGAGCCGAGGCGGCAAGCGGCGAUGAAUGU --.........((((((..-(((((.(((((((((((((..((.----.....----))))))..(((......)))......)))))))))((.....))))))).)))))) ( -37.10) >DroSim_CAF1 14477 102 - 1 --UUCUGUUUCGCAUUUUU-UCGCCACUUGGCUCAGGCUUCAGG----CAGGA----CUAGCCAUUCAUUCAUUUGGUUUUUAUGAGCCGAGGCGGCAAGCGGCGAUGAAUGU --.........((((((..-(((((.(((((((((((((..((.----.....----))))))..(((......)))......)))))))))((.....))))))).)))))) ( -37.10) >DroEre_CAF1 17502 105 - 1 --UUCUGUUUCGCAUUUUU-UCGCCACUUGCCUC-GGCUUCAGC----CAGCAGAAGCCAGCCACUCAUUCAUUUGGUUUUUAUGAGCCGAGGCGGCAAGCGGCGAUGAAUGU --.........((((((..-(((((.((((((..-((((((...----.....)))))).(((..........(((((((....)))))))))))))))).))))).)))))) ( -44.40) >DroYak_CAF1 15451 110 - 1 GUUUCUGUUUCGCAUUUUU-UCGCCACUUGCCUC-GACUUCAGCAGGCCAGGAGUAG-AAGCCAUUCAUUCAUUUGGUUUUUAUGAGCCGAGGCGGCGAGUGGCGAUGAAUGU ...........((((((..-((((((((((((..-(....).....(((.((.((((-((((((..........))))))))))...))..))))))))))))))).)))))) ( -45.80) >DroAna_CAF1 7755 103 - 1 --UUCUGUUUCGCAUUUUUUUCGCAGUUUUGC---AGCUUCUGG----CAUAGAGAG-AGAGCAGCCAUUCAUUUGGUUUUUAUGAGCCGGGGCAGCAAGUGGCGAUGAAUGU --.........((((((...((((...(((((---.(((.((((----(.((...((-(((.(((........))).))))).)).)))))))).)))))..)))).)))))) ( -31.40) >consensus __UUCUGUUUCGCAUUUUU_UCGCCACUUGGCUC_GGCUUCAGG____CAGGA____CAAGCCAUUCAUUCAUUUGGUUUUUAUGAGCCGAGGCGGCAAGCGGCGAUGAAUGU ...........((((((...(((((.((((.(...((((....................)))).........((((((((....))))))))..).)))).))))).)))))) (-24.47 = -25.08 + 0.61)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:06:02 2006